Figure 6.
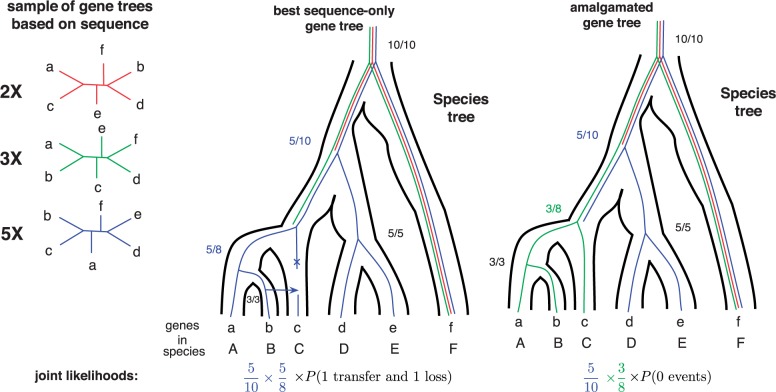
Based on gene trees sampled according to their posterior probability, conditional clade probabilities (CCP) can used to estimate the posterior probability of any tree that can be amalgamated (Höhna and Drummond 2012) from clades present in the sample. Conditional clade frequencies can be used to approximate CCPs and are computed as the fraction of times a particular split of a clade, for example (abc,de) is observed among all trees in which the containing clade, for example (abcde) is found. Estimates based on the sample of trees on the left are shown as fractions for two different gene trees that can be amalgamated. The estimate for a gene tree is given by the product of the frequencies. Amalgamated likelihood estimation (ALE (Szöllősi et al. 2013a)) is a probabilistic approach to exhaustively explore all reconciled gene trees that can be amalgamated as a combination of clades observed in a sample of gene trees. Based on the sample on the left, the tree with the highest posterior probability is the third tree (blue online). Reconciling it with the species tree requires 1 transfer and 1 loss event. It is, however, possible to combine clades present in the second (green online) and third (blue online) trees to produce a gene tree that is not present in the original sample but is identical to the species tree, that is it requires 0 events to draw it into the species tree. Depending on the relative probabilities of and , the joint conditional probability may prefer the scenario without transfer.