Fig 3.
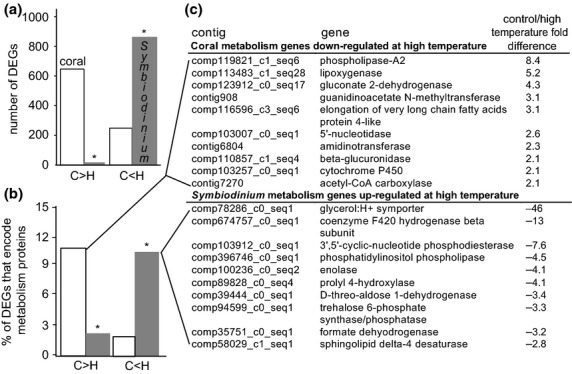
Differentially expressed genes (DEGs) across temperature treatments for each compartment of the Pocillopora damicornis–Symbiodinium endosymbiosis, with an emphasis on metabolism-targeted genes. Differentially expressed genes (DEGs P < 0.01) were sorted into those that were expressed at higher levels in controls (‘C>H’) and those expressed at higher levels in the high-temperature samples (‘C<H’) for both coral (white column) and Symbiodinium (grey column) compartmentss (a). Then, the fraction of the DEG pool from each compartment that was comprised of metabolism genes was calculated (#metabolism DEGs/total number of DEGs × 100) for each compartment for genes differing by twofold or greater between treatments (at either or both sampling times) (b). A random selection of the metabolism-targeted genes that were down-regulated at high temperature at twofold or greater levels in the host coral and up-regulated at twofold or greater levels at high temperature in the Symbiodinium compartment can be found in the embedded table (c). When differences between compartments within each comparison group were determined to be statistically significant by student's t-tests and two-sample proportion tests (P < 0.01) for (a) and (b), respectively, an asterisk (‘*’) has been placed above one of the two columns.
