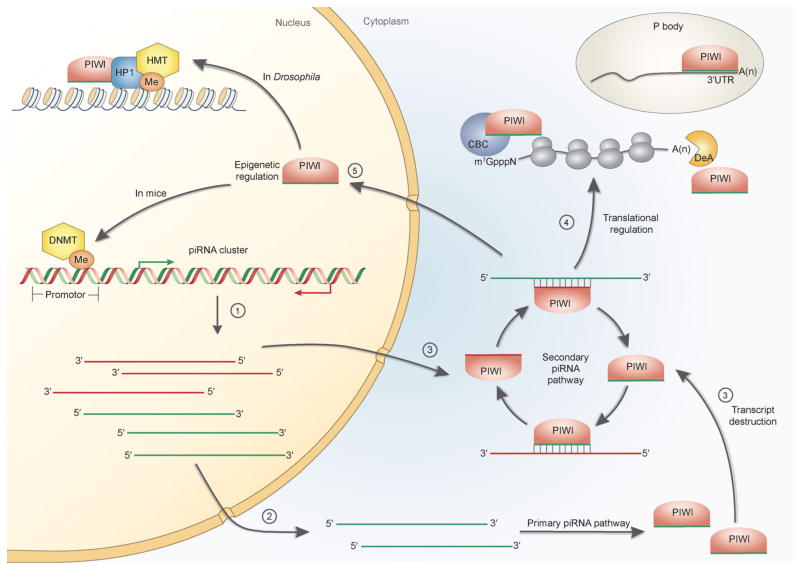
Figure 2.
Known gene regulation mechanisms mediated by the PIWI–piRNA pathway. PIWI proteins and piRNAs regulate the expression of genes and transposon at both transcriptional and post-transcriptional levels. (1) Sense and antisense piRNA precursor transcripts are produced in the nucleus. (2) Antisense piRNA precursor transcripts are transported to the cytoplasm and processed by the primary biogenesis pathway to generate mature sense piRNAs that associate with PIWI proteins. (3) The PIWI–antisense piRNA complexes mediate cleavage of sense piRNA precursors and transposon (and protein-coding) transcripts, which silences transposon and gene expression at the post-transcriptional level. The resulting sense transcripts are taken up by PIWI proteins responsible for sense piRNA binding. The PIWI–sense piRNA complexes then cleave antisense piRNA precursor transcripts to amplify the piRNA biogenesis cycle [48,50]. (4) The PIWI–piRNA complexes are involved in translational regulations by interacting with polysomes, mRNA cap-binding complex (CBC, in mice) [7,74,94], and mRNA deadenylase (DeA, in Drosophila) [133]. In addition, the PIWI–piRNA complexes are associated with P-body components [37,109,136,137] and piRNAs are mapped to the 3′UTR of mRNAs [131,132]. (5) The PIWI–piRNA complexes can enter the nucleus and regulate gene transcription through epigenetic mechanisms including heterochromatin formation [9,11,114–117] and DNA methylation [10,120] in the promoter region of target genes. 3′UTR, 3′ untranslated region; CBC, cap-binding complex; DeA, deadenylase; DNMT, DNA methyltransferase; HMT, histone methyltransferase; HP1, heterochromatin protein 1; Me, methylation.