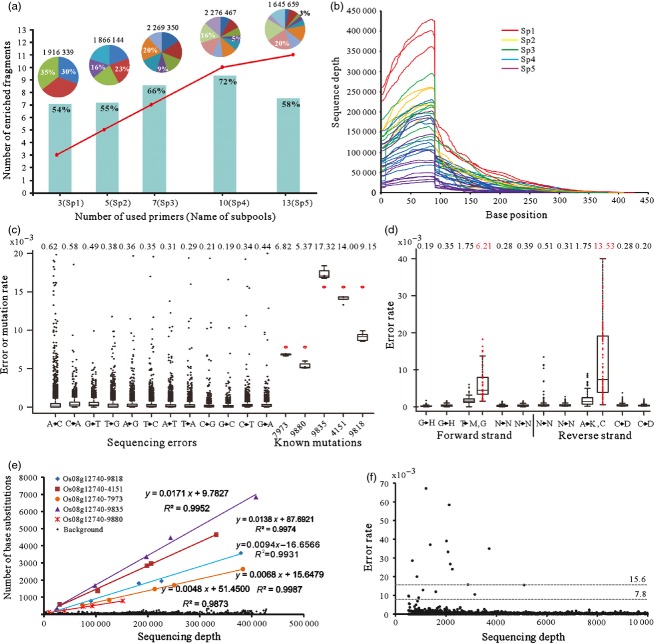
Figure 2.
Summary of the pilot experiment. (a) The number of enriched target fragments and the enrichment efficiency. The pie chart at the top shows the total number of reads and the maximum and minimum proportions of reads in the five enriched libraries produced by varying the number of target-specific semi-nested PCR primers. The red line indicates the number of enriched target fragments when 3–13 target-specific semi-nested PCR primers were used to amplify templates of the corresponding subpools. The histogram in cyan indicates the ratio of on-target sequences. (b) Sequencing depth of the enriched target DNA fragments. (c) A box plot showing the rate of both sequencing errors and genuine mutations in the five subpools. The mean rate of each nucleotide substitution type is shown above the box plot. (d) The sequencing error rate in the vicinity of GGT/ACC trinucleotides. (e) The detected base substitution rates of C to T or G to A plotted against sequencing depth. (f) The background sequencing error rate plotted against sequencing depth. The dotted lines indicate expected allele frequencies of 15.6 × 10−3 and 7.8 × 10−3 in a pooled sample comprising, respectively, a single homozygous and a single heterozygous mutant along with 63 wild-type individuals.