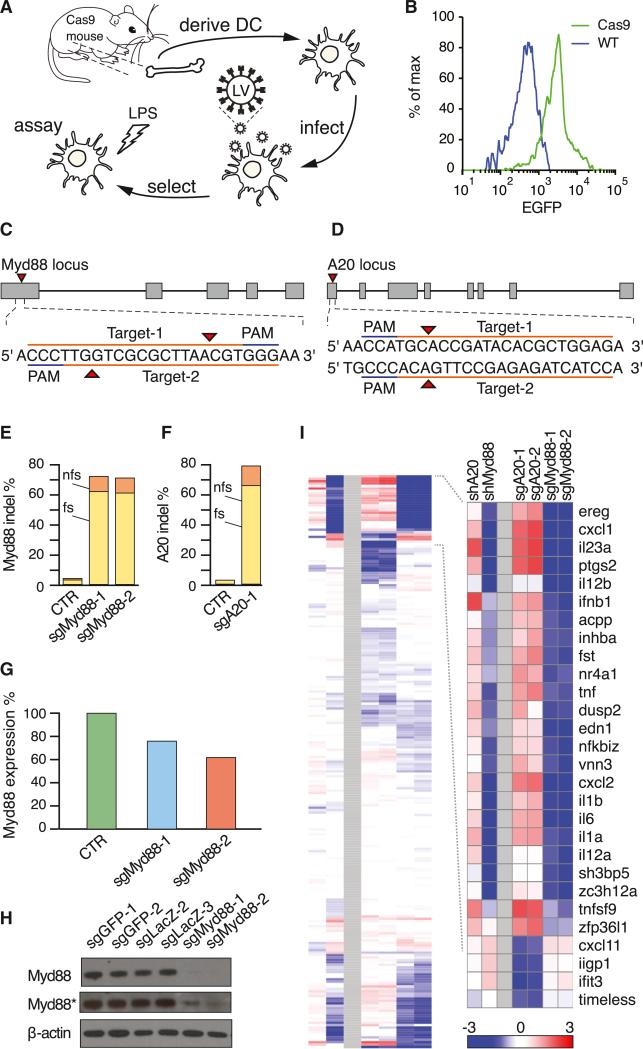
Figure 2. Ex Vivo Genome Editing of Primary Immune Cells Derived from Constitutive Cas9-Expressing Mice.
(A) Schematic of ex vivo genome editing experimental flow.
(B) Flow cytometry histogram of bone marrow cells from constitutive Cas9-expressing (green) and wild-type (blue) mice, showing Cas9-P2A-EGFP expression only in Cas9 mice. Data are plotted as a percentage of the total number of cells.
(C) sgRNA design for targeting the mouse Myd88 locus.
(D) sgRNA design for targeting the mouse A20 locus.
(E) Myd88 indel analysis of constitutive Cas9-expressing DCs transduced with either a Myd88-targeting sgRNA (sgMyd88-1 and sgMyd88-2) or controls (CTR, average of four control sgRNAs), showing indel formation only in Myd88-targeted cells. Data are plotted as the percent of Illumina sequencing reads containing indels at the target site. Mutations are categorized as frameshift (fs, yellow bar) or non-frameshift (nfs, orange bar).
(F) A20 indel analysis of constitutive Cas9-expressing DCs transduced with either an A20-targeting sgRNA (sgA20-1) or controls (CTR, average of four control sgRNAs), showing indel formation only in A20-targeted cells. Data are plotted as the percent of Illumina sequencing reads containing indels at the target site. Mutations are categorized as frameshift (fs, yellow bar) or non-frameshift (nfs, orange bar).
(G) Myd88 mRNA quantification of constitutive Cas9-expressing DCs transduced with either Myd88-targeting sgRNA (sgMyd88-1 or sgMyd88-2) or controls (CTR, average of six control sgRNAs), showing reduced expression only in Myd88-targeted cells. Data are plotted as Myd88 mRNA levels from Nanostring nCounter analysis.
(H) Immunoblot of constitutive Cas9-expressing DCs transduced with either Myd88-targeting sgRNA (sgMyd88-1 or sgMyd88-2) or controls (four control sgRNAs), showing depletion of Myd88 protein only in Myd88-targeted cells. β-actin was used as a loading control. (Asterisk) Overexposed, repeated-measurement.
(I) Nanostring nCounter analysis of constitutive Cas9-expressing DCs transduced with either Myd88-targeting sgRNA (sgMyd88-1 or sgMyd88-2) or shRNA (shMyd88), A20-targeting sgRNA (sgA20-1 or sgA20-2), or shRNA (shA20), showing an altered LPS response. (Inset) The cluster showing the highest difference between Myd88- and A20-targeting sgRNAs, including key inflammatory genes (IL1a, IL1b, Cxcl1, Tnf, etc.). (Red) High; (blue) low; (white) unchanged; based on fold change relative to measurements with six control sgRNAs. See also Figure S2.