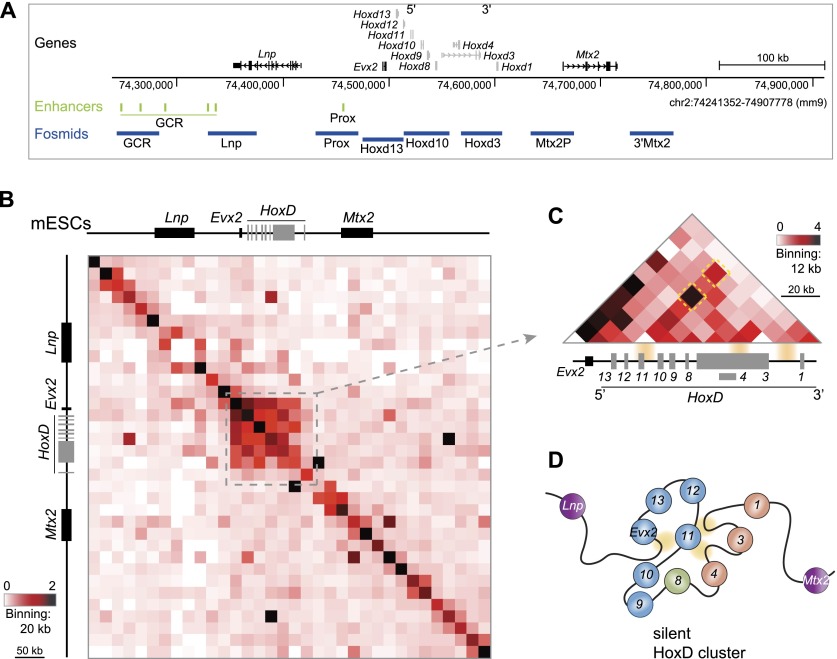
Figure 1.
5C analysis of HoxD in ESCs. (A) A 670-kb region analyzed by 5C in OS25 mESCs, encompassing Mtx2, Hoxd genes, Evx2, and Lnp. Distal regulatory elements (Montavon et al. 2011) are highlighted in green. Positions of FISH probes are indicated in blue. Genome coordinates are from the NCBI37/mm9 assembly of the mouse genome. (B) Analysis of chromatin organization in undifferentiated mESCs by 5C sequencing across the 670-kb region shown in A. The heat map shows 5C data binned over 20-kb windows. Heat map intensities represent the average of interaction frequency for each window, color-coded according to the scale shown. Data for a biological replicate are in Supplemental Figure S1A. Unprocessed normalized data are shown in Supplemental Figure S7. All interaction frequencies were first normalized based on the total number of sequence reads in the 5C data set. (C) High-resolution (12-kb binning) zoomed-in view of the 5C data over Exv2 and the HoxD locus. The two contacts conserved in human embryonic carcinoma cells are indicated with dashed yellow boxes. (D) Two-dimensional schematic interpretation of the 5C data in OS25 mESCs illustrating the folded nature of the Evx2–HoxD domain (not to scale). Contacts between Hoxd11 and regions downstream from d1, d3, and Evx2 are highlighted in yellow.