Figure 6.
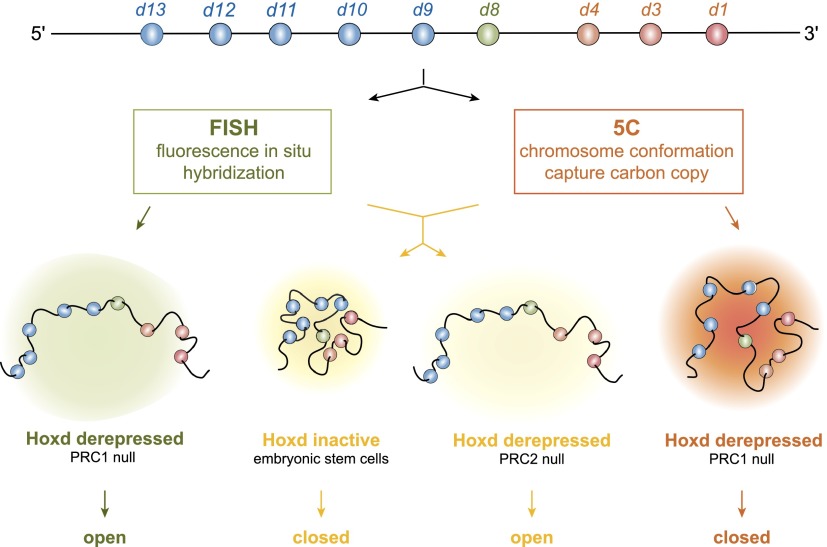
FISH and 5C analysis can yield compatible or discordant chromatin topographies at high resolution. (Top) Schematic of the HoxD locus showing the 5′ (blue) and 3′ (orange) Hoxd genes. (Bottom) The views of chromatin topography for HoxD extrapolated from FISH or 5C data are shown for wild-type and polycomb mutant ESCs. (Middle) For wild-type and PRC2-null cells, FISH and 5C give coherent views of a compact (wild-type) versus unfolded (PRC2-null) chromatin conformation. However, in the case of PRC1-null cells, FISH (left) indicates an unfolded chromatin conformation similar to that seen for PRC2-null cells, whereas 5C (right) suggests a much more tightly folded domain.