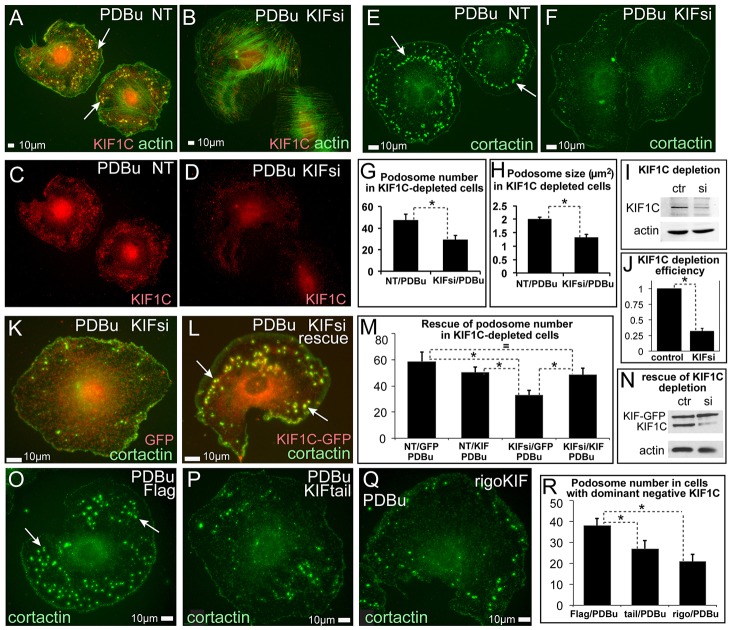
Fig. 2.
Podosome formation in A7r5 cells depends on KIF1C. (A–F) Immunofluorescence visualization of podosomes by actin (phalloidin, green, A,B) and cortactin (green, E,F). KIF1C (red) is shown in C,D for cells in A,B. NT, non-targeted control siRNA-treated; KIFsi, KIF1C-depleted. (B,D,F) After KIF1C depletion only few immature podosomes are detected. The remaining KIF1C is detected in the cell center (D). (G) Podosome numbers based on data similar to that shown in E,F. Data show the mean+s.e.m. (N = 32); *P<0.01 (Student's unpaired two-tailed t-test). (H) Average of mean podosome area per cell (µm2) based on data similar to that shown in E,F. Data show the mean+s.e.m. (N = 32). *P<1×10−6 (Student's unpaired two-tailed t-test). (I) Western blotting indicates significant depletion of KIF1C. Actin is shown as a loading control. Ctr, control; si, KIF1C-depleted. (J) Quantification of KIF1C levels detected by western blotting. Data show the mean+s.e.m. (N = 3); *P<0.001 (Student's unpaired two-tailed t-test). (K,L) Re-expression of KIF1C–GFP (red) in KIF1C-depleted cells (L) rescues podosome formation as compared with that of GFP-expressing KIF1C-depleted cells (K). GFP is pseudo-colored red. Cortactin, green. (M) Podosome numbers in KIF1C-depleted and rescued cells. Data show the mean+s.e.m. (N = 45); *P<0.05 (Student's unpaired two-tailed t-test). (N) Western blotting indicates KIF1C–GFP expression in control and KIF1C-depleted cells. Actin is shown as a loading control. (O–Q) Expression of dominant-negative FLAG-tagged KIF1C cargo-binding domain (P) or rigor motor mutant (Q) suppresses podosome formation as compared with that of controls (O). Cortactin, green. All arrows indicate podosomes. (R) Podosome numbers in cells expressing dominant-negative constructs. Data show the mean+s.e.m. (N = 45); *P<0.05 (Student's unpaired two-tailed t-test).