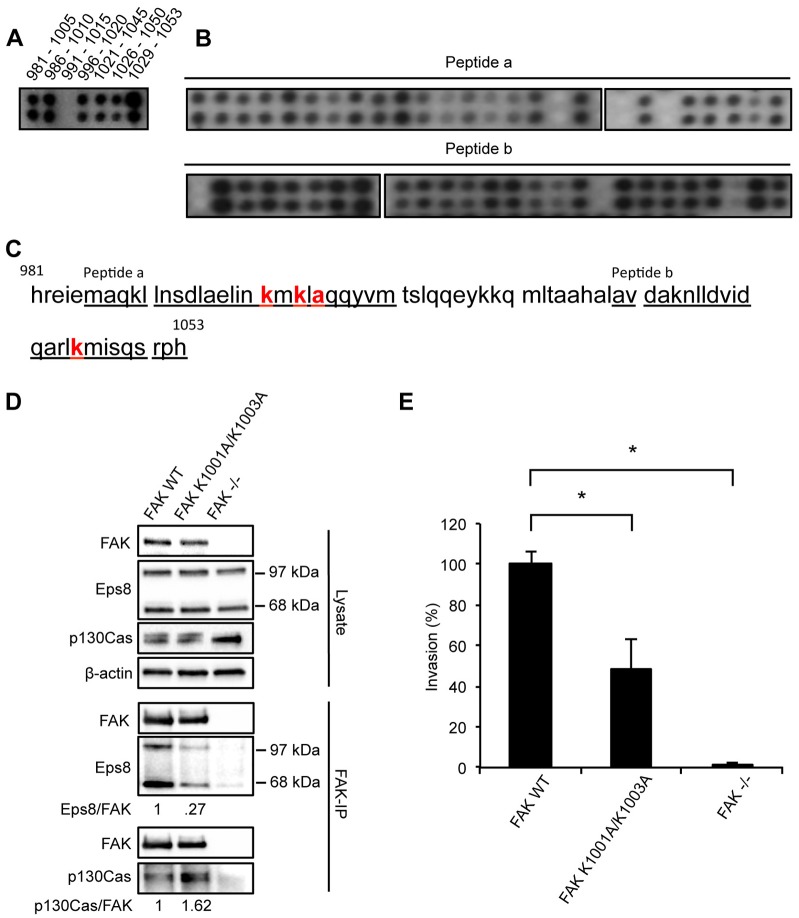
Fig. 3.
Eps8 binds to the C-terminus of FAK. (A–C) Identification of the Eps8-binding sequence in FAK. (A) Overlapping 25-mer peptides spanning the C-terminus of FAK (amino acids 981–1053) were spotted onto nitrocellulose, incubated with recombinant Eps8 and probed with anti-Eps8 antibody. (B) The core binding sequences in peptide a (top panel) and peptide b (bottom panel) were identified by alanine scanning where one amino acid was mutated at a time. (C) The sequences of the 25-mer peptides (peptide a and peptide b) used for the alanine scanning are underlined and the identified core binding sites is marked in bold red. (D) FAK was immunoprecipitated (IP) from FAK WT, FAK K1001A/K1003A and FAK−/− cell lysates using anti-FAK antibody conjugated to agarose (clone 4.47), followed by western blot analysis with anti-FAK, anti-Eps8 and anti-p130Cas antibodies (lower panels). Eps8 immunoblotting detects both known isoforms with molecular masses of 97 kDa and 68 kDa. Anti-β-actin antibody was used as a loading control. (E) Invasion assay. SCC FAK WT, FAK K1001A/K1003A and FAK−/− cells were seeded on growth factor reduced Matrigel in serum-free conditions. Invasion towards serum gradient was visualized after 72 h by staining the cells with calcein. Results are mean±s.e.m. *P≤0.01 (Student's t-test). Quantification is representative of three independent experiments.