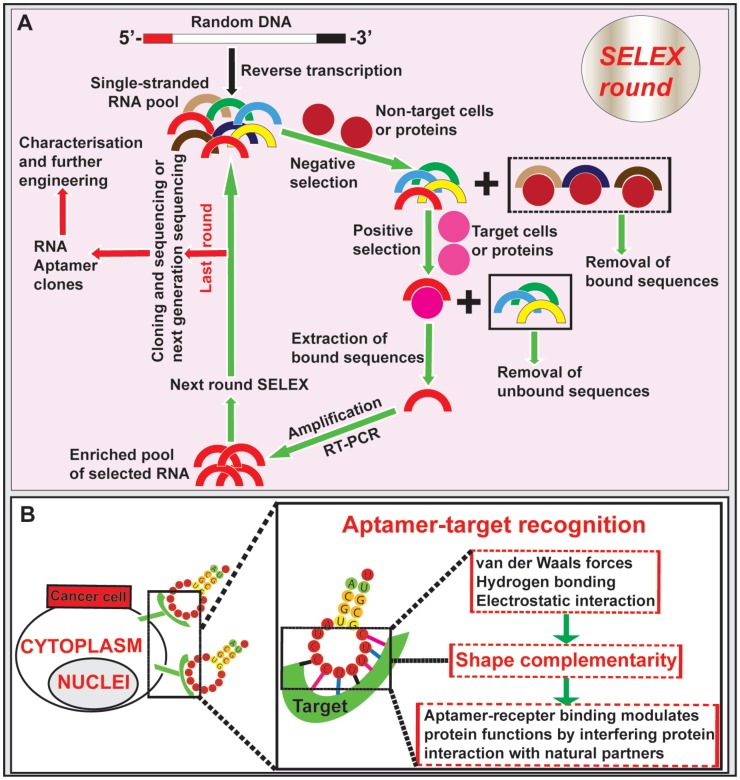
Figure 1.
Isolation of aptamer and aptamer-target cell interaction. (A) Schematic illustration of RNA aptamer generation by SELEX. Negative control cells or proteins are incubated with a large pool (> 1 x 1014) of single-stranded RNA oligonucleotides with random sequences, followed by removal of bound RNAs. The unbound sequences are then subjected to positive selection by incubating with specific targets of cells or proteins. The bound aptamers are eluted and amplified by RT-PCR, followed by new SELEX rounds. After the last round of selection (typically 6-10 round), the remaining RNA aptamers are converted to DNA and subjected to next generation sequencing or conventional cloning followed by characterization and further engineering. (B) Schematic diagram depicting the aptamer-target interactions. Aptamers bind to targets at the cell surface through shape complementarity involving non-covalent bonds.