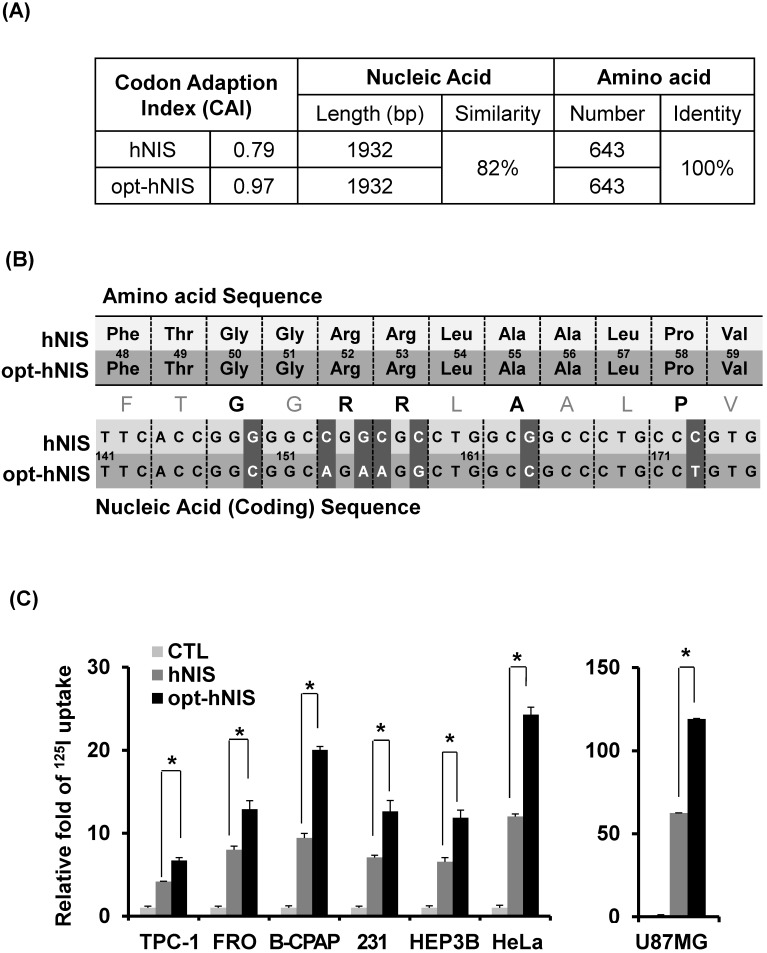
Figure 1.
Comparison between the wild-type human NIS (hNIS) and codon-optimized (opt-hNIS) gene. (A) The predictive level of gene expression by tRNA abundance in humans is described by the Codon Adaptation Index (CAI). The maximum possible CAI is set to 1.0, corresponding to the quality value of the most frequently used codon for a given amino acid at the desired expression. Despite differences in coding sequence, the amino acids produced are identical. (B) Sequence comparison of a specific portion of the hNIS and opt-hNIS genes, i.e., amino acid sequence 48 to 59 (FTGGRRLAALPV) corresponding to nucleic acid sequence 141 to 176 bp. Bold characters in the amino acid sequence indicate amino acids with their codons changed after optimization. Dark gray blocks in the nucleic acid sequences indicate the location of the changed codons having high frequency in humans by optimization. (C) Increased radioiodine uptake in cancer cells by hNIS and opt-hNIS transfection. Various cancer cells such as thyroid (FRO, TPC-1, B-CPAP), breast (MDA-MB-231), liver (Hep3B), cervical (HeLa), and glioma (U87MG) were transiently transfected with pcDNA3.1/hNIS or opt-hNIS to evaluate functional changes of hNIS expression (P < 0.01, N=3).