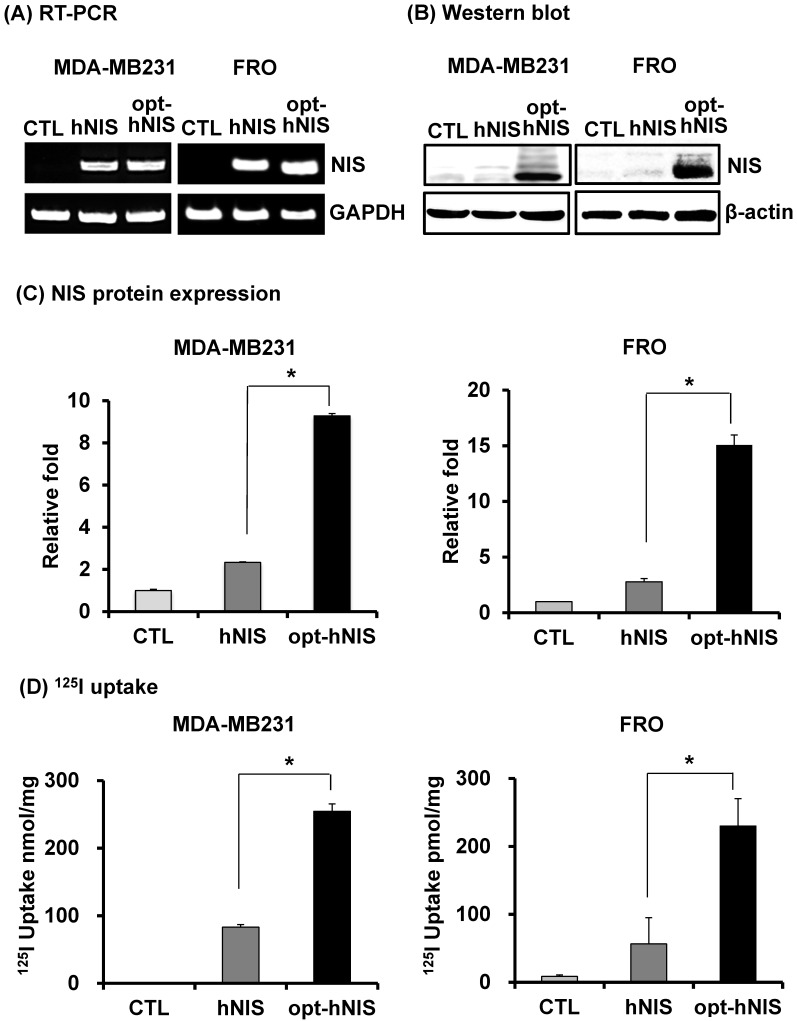
Figure 2.
Comparative analysis of NIS expression between cells expressing hNIS or opt-hNIS genes. (A) RT-PCR analysis of NIS transcripts in hNIS-/or opt-hNIS-expressing cells (MDA-MB-231 and FRO cells) established from monoclonal selections of retrovirus-mediated stably transduced cells. GAPDH was used as an internal control. Marked overexpression of hNIS mRNA was observed in both hNIS- and opt-hNIS-expressing cells. No expression was observed in the parental cells (CTL). (B) Western blot analysis of NIS protein in hNIS- or opt-hNIS-expressing cells. Multiple bands of glycosylated NIS were observed in both hNIS- and opt-hNIS-expressing cells, but more NIS protein was produced in opt-hNIS-expressing cells. (C) Quantitative analysis of protein expression in hNIS- or opt-hNIS-expressing cells. (D) 125I uptake assay in hNIS- or opt-hNIS-expressing cells. All experiments were performed in triplicate, and bars represent means ±SD (*, P<0.001).