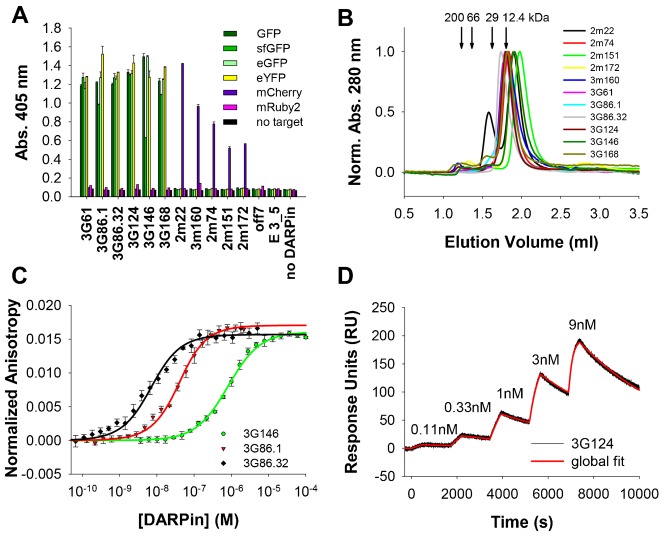
Fig. 1. Specificity, oligomeric state and affinity of anti-GFP and anti-mCherry DARPins.
(A) ELISA experiments show a high specificity of selected DARPins towards their cognate target and closely related proteins. Off7 is a control DARPin binding to maltose binding protein (MBP), E 3_5 is an unselected DARPin. All bars represent mean values of duplicates, error bars represent standard deviations. (B) Analysis of the oligomeric state by SEC shows that most DARPins are predominantly monomeric. Due to low extinction coefficients at 280 nm for the chromatograms of 2m22 and 2m74 the absorption at 230 nm is shown. Arrows indicate the elution volumes of the molecular weight standard with the respective MWs. (C) Example of FA assays of different DARPins binding to sfGFP. The solid line indicates a fit to a 1:1 binding model. Extracted KDs for all DARPins can be found in Table 1. (D) Example of a kinetic titration SPR experiment of 3G124 binding to GFP. The concentrations of the five DARPin injections are indicated in the graph. Fit to a global 1:1 kinetic titration binding model is indicated in red. Extracted association and dissociation rates and KDs for several DARPins are summarized in Table 1; additional sensograms are shown in supplementary material Fig. S4.