Figure 3.
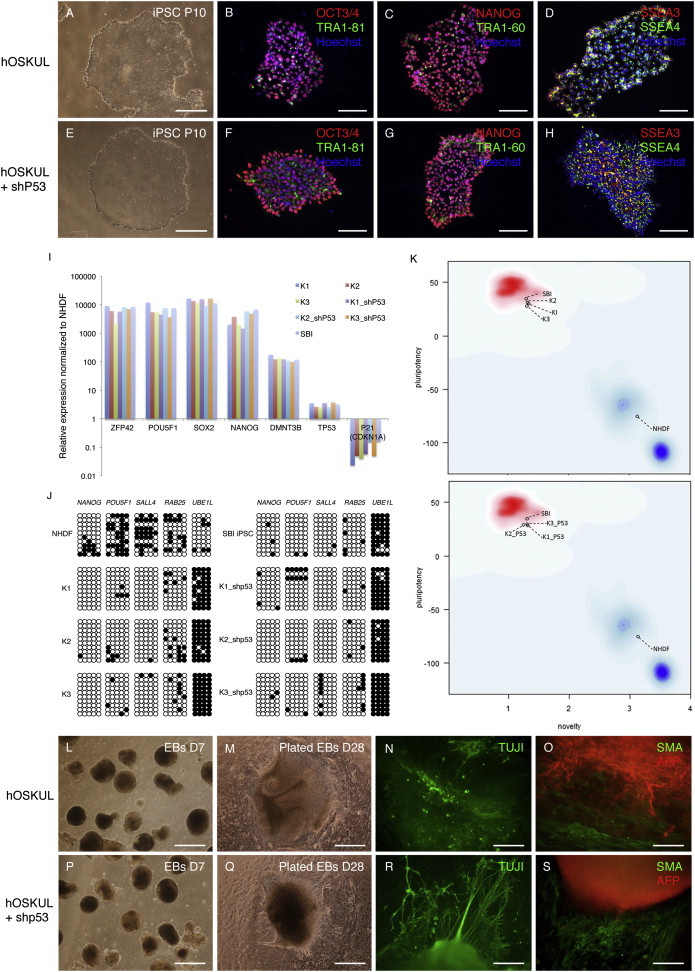
iPSCs Generated by Transient p53 Suppression Display Normal In Vitro Characteristics
(A–H) (A and E) Phase contrast morphology and (B–H) immunocytochemical staining of induced pluripotent stem cells (iPSCs) generated with or without (w/wo) a short hairpin to p53 (shp53) with the pluripotency markers OCT3/4, TRA1-81, and Hoechst (B and F), NANOG, TRA1-60, and Hoechst (C and G), and SSEA3, SSEA4, and Hoechst (D and H).
(I) Quantitative real-time PCR with the pluripotency markers ZFP42, POU5F1 (OCT4), SOX2, NANOG, DNMT3B, and TP53 and P21. Relative expression is shown as the fold change (2−ΔΔCt) with normal human dermal fibroblasts (NHDFs) as reference. A commercial iPSC line (SBI) was included.
(J) Schematic view of the methylation state of selected CpG sites in POU5F1 (OCT4), SAL4, RAB25, and UBE1L. Each row represents a separate clone and each circle represents a CpG site. Open circles represent unmethylated sites and black circles represent methylated sites.
(K) PluriTest algorithm results of transcriptome profiles including a reference iPSC line (SBI) and NHDFs. The PluriTest results are plotted in a density distribution for previously referenced pluripotent stem cells (red cloud) and somatic cells (blue cloud).
(L–S) (L, M, P, and Q) Morphology of embryoid bodies (EBs) on day 7 (L and P), plated EBs on day 28 (M and Q), and immunocytochemical staining of plated EBs on day 28 with TUJI (N and R), smooth muscle actin (SMA), and alpha-fetoprotein (AFP) (O and S).
Scale bars, 400 μm (A, E, L, M, P, and Q), 100 μm (B–D and F–H), or 200 μm (N, O, R, and S). See also Figure S3 and Tables S1–S3.
