Abstract
Sulfate-reducing bacteria (SRB) can interact syntrophically with other community members in the absence of sulfate, and interactions with hydrogen-consuming methanogens are beneficial when these archaea consume potentially inhibitory H2 produced by the SRB. A dual continuous culture approach was used to characterize population structure within a syntrophic biofilm formed by the SRB Desulfovibrio vulgaris Hildenborough and the methanogenic archaeum Methanococcus maripaludis. Under the tested conditions, monocultures of D. vulgaris formed thin, stable biofilms, but monoculture M. maripaludis did not. Microscopy of intact syntrophic biofilm confirmed that D. vulgaris formed a scaffold for the biofilm, while intermediate and steady-state images revealed that M. maripaludis joined the biofilm later, likely in response to H2 produced by the SRB. Close interactions in structured biofilm allowed efficient transfer of H2 to M. maripaludis, and H2 was only detected in cocultures with a mutant SRB that was deficient in biofilm formation (ΔpilA). M. maripaludis produced more carbohydrate (uronic acid, hexose, and pentose) as a monoculture compared to total coculture biofilm, and this suggested an altered carbon flux during syntrophy. The syntrophic biofilm was structured into ridges (∼300 × 50 μm) and models predicted lactate limitation at ∼50 μm biofilm depth. The biofilm had structure that likely facilitated mass transfer of H2 and lactate, yet maximized biomass with a more even population composition (number of each organism) when compared to the bulk-phase community. Total biomass protein was equivalent in lactate-limited and lactate-excess conditions when a biofilm was present, but in the absence of biofilm, total biomass protein was significantly reduced. The results suggest that multispecies biofilms create an environment conducive to resource sharing, resulting in increased biomass retention, or carrying capacity, for cooperative populations.
Keywords: anaerobic, carrying capacity, hydrogen transfer, population intermixing, sulfate-reducing bacteria
INTRODUCTION
Symbiosis (“living together”) and specifically mutualism, whereby both parties incur a benefit from living together, is widespread throughout the biosphere with well-studied examples in and across all three domains of life (Yeoh et al., 1968; Boucher, 1988; Kato et al., 2011; Moissl-Eichinger and Huber, 2011; Plugge et al., 2011; Gokhale and Traulsen, 2012; Sieber et al., 2012). In communities of bacteria and archaea, mutualism is typically referred to as syntrophy (“eating together”) where by-products of one metabolism serve as substrates for another metabolism (Sieber et al., 2012). The syntrophy between sulfate-reducing bacteria (SRB) and methanogenic archaea is of interest because these guilds both play crucial roles in many different anaerobic environments. SRB link the carbon, sulfur, and nitrogen biogeochemical cycles via carbon-oxidation and sulfate reduction (Purdy et al., 2002; Canfield et al., 2010) and also contribute to redox gradients of microbial ecosystems via the production of sulfide compounds (Moreau et al., 2010; Xie et al., 2013). Methanogenic archaea are responsible for the three largest sources of methane flux to the atmosphere (wetlands, ruminants, and rice cultivation) and form the basis of most anaerobic environments (natural and man-made) that convert CO2, H2, and/or acetate/methyl-groups to methane (Thauer et al., 2008; Neef et al., 2010; van Groenigen et al., 2012; Schlesinger and Bernhardt, 2013).
The nature of SRB-methanogen interactions is complex and fluctuates based on substrate flux and availability (Leloup et al., 2009; Stams and Plugge, 2009; Plugge et al., 2011). In the presence of sulfate, methanogens are typically outcompeted by SRB using H2, formate, and acetate as electron donors for sulfate reduction (Plugge et al., 2011). SRB can alternatively form mutualistic partnerships with hydrogenotrophic methanogens in the absence of sulfate by proton reduction to form H2 gas. The reaction is kept favorable when hydrogenotrophic methanogens consume H2, keeping the partial pressure low and thereby eliminating the inhibitory effect of this end-product on the SRB (Figure S1A; McInerney et al., 2009; Stams and Plugge, 2009). Inhabitants of anaerobic ecosystems are assumed to function at the thermodynamic limit for energy generation and biomass production given system constraints (Bryant et al., 1977; Thauer et al., 2008; McInerney et al., 2009; Kato and Watanabe, 2010). When one metabolism is obligately coupled to another through interspecies H2, formate, or electron transfer, organisms must persist by sharing the overall free energy of the reaction (Kato and Watanabe, 2010). Therefore, syntrophic physiology plays an important role in microbial communities dominated by fluctuations in nutrient availability and stress, where community interactions are thought to provide stability (Hansen et al., 2007).
It is well-accepted that microorganisms can exist attached to surfaces and each other, often surrounded by extracellular polymeric substances (EPS; Hall-Stoodley et al., 2004; Gross et al., 2007; Stewart and Franklin, 2008). Biofilms have been described from environments where there are liquid–solid or liquid–gas interfaces that include terrestrial and deep-sea hydrothermal features, riparian zones, ship hulls, metal pipes, saturated soils, and the human body (Hall-Stoodley et al., 2004), but much of the work to identify the driving force and genetic control over biofilm formation has been done with pure culture studies (e.g., Gross et al., 2007; Stewart and Franklin, 2008; Perez-Osorio et al., 2010; Clark et al., 2012).
The structure and function of multispecies biofilms can be more complex than monocultures, and biofilm structure from several environments has been characterized with confocal scanning laser microscopy (CSLM) using fluorescence in situ hybridization (FISH) and immunofluorescence (Møller et al., 1998; Hall-Stoodley et al., 2006; Al-Ahmad et al., 2009; Stams and Plugge, 2009; Jakubovics, 2010; Zijnge et al., 2010). We have recently shown that the structure of a mixed biofilm community is dependent upon the nature of the interactions (i.e., cooperative or competitive) and that the degree of intermixing of two-member communities is greater during cooperation versus competition (Momeni et al., 2013). The dependence of biofilm structure on function has also been demonstrated in a dual culture system where commensal biofilm cells interacted closely in mixed micro-colonies, while non-commensals formed separate non-interacting biofilm micro-colonies (Nielsen et al., 2000). Despite the ubiquity of biofilms and importance of anaerobes, little work has been done to understand how biofilm structure affects function in anaerobic microbial communities (Raskin et al., 1996; Nielsen et al., 2000; Brenner and Arnold, 2011; Bernstein et al., 2012).
While interactions between SRB and methanogens have been studied, very little has been done to characterize the emergent properties of interactive populations in anaerobic biofilms. The purpose of this work was to characterize the relationship between biofilm structure and function in biofilm formed by a SRB and a hydrogenotrophic methanogen cultured syntrophically under nutrient limitation and nutrient excess conditions (i.e., carbon source and electron donor/acceptor). We hypothesized that a biofilm would be functionally more efficient in terms of product formation (i.e., CH4) compared to populations in the bulk aqueous phase. A system was developed for anaerobic continuous culture where biofilm and planktonic growth phases could be monitored to determine the difference in biomass yield per mass flux of lactate and methane under varying conditions. To compare the biomass yield of biofilm to the biomass yield of planktonic cocultures, we removed the biofilm from a series of reactors, and in another experiment, used a biofilm deficient mutant coculture.
MATERIALS AND METHODS
CULTURE CONDITIONS
Desulfovibrio vulgaris ATCC 29579 and Methanococcus maripaludis S2 (DSM 14266) were continuously cultured in modified 1L CDC reactors (BioSurface Technologies Corp., Bozeman, MT, USA) for anaerobic biofilm growth (Figure S1B). Biofilm coupon holders were modified to hold glass microscope slides cut to 7.6 cm × 1.8 cm as previously described (Clark et al., 2012). Both monocultures and cocultures were grown in coculture medium (CCM), a bicarbonate buffered, basal salts medium without choline chloride (Walker et al., 2009). Monoculture D. vulgaris medium was supplemented with 25 mM sodium sulfate, or grown in standard lactate-sulfate medium (LS4D) with 30 mM lactate and 25 mM sodium sulfate as previously described Clark et al. (2006). Headspace (290 mL) was purged at 20 mL/min with anoxic 80% N2:20% CO2 (v/v) for coculture and monoculture D. vulgaris or 80% H2:20% CO2 for monoculture M. maripaludis through a 0–20 SCCM mass controller (Alicat Scientific, Tucson, AZ, USA). Reactors were maintained at 30°C with stirring at 80 rpm. The reactor aqueous phase (375 mL) was inoculated with 20 mL of mid-exponential phase planktonic cultures grown from glycerol freezer (–80°C) stocks in 40 mL of CCM in 125 mL serum bottles. Fresh CCM in a 20 L glass carboy was continuously sparged with sterile anoxic 80% N2:20% CO2 and supplied at a dilution rate of 0.017 h-1 starting after 48 h of batch growth by a Masterflex L/S pump (Cole-Parmer Instruments Co., Vernon Hills, IL, USA). Batch monoculture M. maripaludis was grown in Balch tubes in 5 mL of CCM with 30 mM acetate in lieu of lactate, prepared under 80% N2:20% CO2, and then pressurized after autoclaving to 200 kPa with 80% H2:20% CO2.
GAS CHROMATOGRAPHY
Gas measurements were made by automated injections (250 ms) of reactor headspace via a 16-port stream selector (Vici-Valco Instruments Co. Inc., Houston, TX, USA) to a 490microGC (Agilent Technologies Inc., Santa Clara, CA, USA) equipped with dual channels and dual thermal conductivity detectors. Molsieve5A and PoraplotQ (both 10 m) columns were run with Helium carrier gas at 145 kPa and 80°C with injectors at 110°C and heated sample line at 40°C. The CDC reactor lids were fitted with stainless steel fittings (Swagelok, Idaho Falls, ID, USA) to accommodate 1/16” PEEK tubing to the stream selector. Scotty calibration gasses were used as standards (Air Liquide America Specialty Gases, Plumsteadville, PA, USA).
FLUORESCENCE IN SITU HYBRIDIZATION
Fluorescence in situ hybridization was used on scraped biofilms to determine relative biovolume of each cell type. Whole biofilm on the glass coupon was fixed in 4% paraformaldehyde in a 50 mL conical tube for 3 h at 4°C, then scraped into a well on a Teflon coated slide (Paul Marienfeld GmbH & Co. KG, Lauda-Königshofen, Germany). Dried biofilm was dehydrated and hybridized in buffer solution containing 180 μL 5 M NaCl, 20 μL 1 M Tris HCl, 449 μL double deionized (dd) H2O, 1 μL 10% SDS and 350 μL deionized formamide (final concentration 35%) with 3 ng each of probes EUB338 (GCT GCC TCC CGT AGG AGT) double labeled with Cy3 and ARCH915 (GTG CTC CCC CGC CAA TTC CT) double labeled with Cy5 for 4 h at 46°C in a humid chamber (Stoecker et al., 2010). Samples were washed in prewarmed washing buffer containing 700 μL 5 M NaCl, 1 mL 1 M TrisHCl, 500 μL 0.5 M EDTA and raised to 50 mL with ddH2O, at 47°C for 10 min, then dipped in ice cold ddH2O and quickly dried with compressed air. Samples were mounted with Citifluor AF1 antifadent (Citifluor Ltd., Leicester, UK) for CLSM. 3D-FISH (Daims et al., 2006) was used to determine colocalization patterns on intact, unscraped biofilm. For 3D-FISH, whole fixed biofilm on the glass coupons was embedded in polyacrylamide prior to dehydration (Daims et al., 2006; Brileya et al., 2014).
CONFOCAL LASER SCANNING MICROSCOPY
Fluorescently labeled biofilm was imaged using a Leica TCS SP5 II inverted confocal laser scanning microscope with 488, 561, and 633 nm lasers and appropriate filter sets for Cy3 and Cy5. Polyacrylamide-embedded whole biofilm for 3D-FISH and fluorescently stained hydrated biofilm were imaged on a Leica TCS SP5 II upright confocal laser scanning microscope using a 63x 0.9 NA long working distance (2.2 mm) water dipping objective (Leica Microsystems, Exton, PA, USA).
5-CYANO-2,3-DITOLYL TETRAZOLIUM CHLORIDE (CTC) STAINING
Biofilm metabolic potential was assessed using the redox stain 5-cyano-2,3-ditolyl tetrazolium chloride (CTC). Whole hydrated biofilm coupons were removed in an anaerobic chamber and incubated in freshly prepared anoxic 0.05% CTC solution for 2 h as in Stewart et al. (1994). The reaction was stopped with 5% formaldehyde and rinsed with ddH2O. Hydrated biofilm was stained with 1 μg/mL DAPI for 20 min in the dark and rinsed with ddH2O before CLSM.
CELL COUNTS AND BIOFILM RELATIVE ABUNDANCE
One milliliter of planktonic phase was fixed in formaldehyde (final concentration 2%) overnight then diluted as necessary and stained for 20 min in the dark with an equal volume of filtered 0.3g/L Acridine Orange. Stained samples were collected through a filter chimney on a black polycarbonate track-etched isopore filter (EMD Millipore Corp., Billerica, MA, USA) and imaged on a Nikon Eclipse E800 microscope with a mercury bulb for fluorescence. At least ten random fields of view were analyzed and cells were counted via integrated morphometry analysis in MetaMorph version 7.6 (Molecular Devices, Sunnyvale, CA, USA).
Biofilm relative abundance (biovolume) was determined using thirty CLSM images per sample, captured from random locations in x, y, and z planes. MetaMorph was used to measure the thresholded area of the two channels in each image.
PROTEIN NORMALIZATION
One M. maripaludis cell and one D. vulgaris cell are not the same shape or volume, so average biomass (protein) per cell was determined using monocultures. Biological duplicates of each monoculture were grown to late exponential phase in 125 mL serum bottles. One portion was filtered and dried to determine dry weight per cell. The Lowry protein assay was done in triplicate on each culture to determine protein biomass weight per volume. Additionally cells were fixed and stained for counting as described above. Twenty fields of view were analyzed for each culture to determine cell number per volume and area per cell using MetaMorph version 7.6 software (Figure S2). Protein per cell area was observed to be equivalent in both cell types on average, so no correction factor was applied when determining the fraction of biofilm biovolume contributed by M. maripaludis and D. vulgaris. Total protein per cell was found to be skewed toward one cell of D. vulgaris containing more protein than one cell of M. maripaludis (Figure S2). Therefore when a total planktonic protein measurement was related to cell counts of each population, a correction factor was also applied where 40% of one protein unit was attributed to M. maripaludis and 60% to D. vulgaris.
1-D BIOFILM ACCUMULATION MODEL
Diffusion in the biofilm was modeled using a biofilm accumulation model (BAM; Wanner and Gujer, 1986) to predict effects of biofilm thickness, inlet substrate concentration, and volumetric flow rate on methane production and cell ratios. Input parameters are listed in Table S1. Rate coefficients for substrates were Ks = 1 while stoichiometric coefficients were 1/yield. Yields were calculated based on Gibbs Free Energies for the associated half reactions normalized to one electron. Aqueous diffusion coefficients (Daq) at 25°C for substrates (Stewart, 2003) were corrected to 30°C using D30/D25 = 1.135. Daq of lactate was calculated as in Wilke and Chang (1955):
Biofilm accumulation model allows for input of a ratio of the effective diffusion coefficient to the aqueous diffusion coefficient (De/Daq) which is then applied to all solutes to account for the decreased diffusion observed in the biofilm matrix compared to water.
ELECTRON MICROSCOPY
Micrographs in Figures 1A,C,D and 3 were collected on a Zeiss Supra55VP FE-SEM. Biofilm was fixed in a solution of 2% paraformaldehyde, 2.5% glutaraldehyde and 0.05 M Na-cacodylate overnight at room temperature. Coupons were rinsed and stepwise dehydrated in ethanol before being cut and critical point dried on a Samdri-795 (Tousimis Research Corporation, Rockville, MD, USA). Glass pieces with dry biofilm were mounted on SEM stubs with double-sided carbon tape and silver, then sputter coated with Iridium for 35 s at 35 mA.
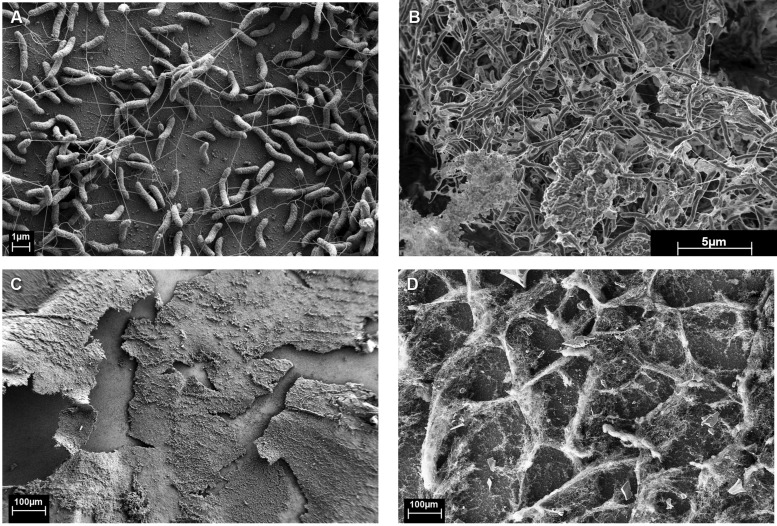
FIGURE 1.
Monocultures of (A,C) Desulfovibrio vulgaris biofilm from continuous culture (5,420X and 101X, respectively). (B,D) Coculture biofilm of D. vulgaris and M. maripaludis (10,000X and 100X, respectively).
FIGURE 3.
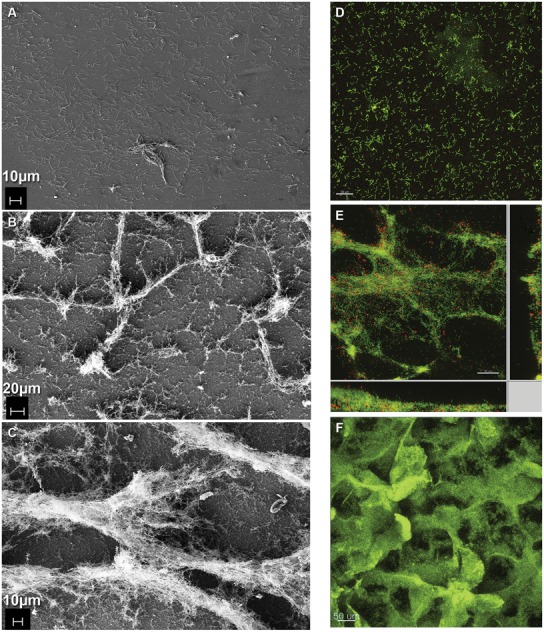
Formation of coculture biofilm over time. (A–C) Electron micrographs of fixed coculture at (A) early (386X, 0 h; B) intermediate (243X, 48 h; C) steady-state (336X, 240 h) time points. (D–F) Fluorescence micrographs of coculture biofilm (D,E) embedded in polyacrylamide and hybridized with domain-specific oligonucleotide probes labeled green for D. vulgaris and red for M. maripaludis at (D) early (0 h) and (E) intermediate (48 h) time points. (F) Intact hydrated biofilm unfixed and stained with Acridine Orange at steady-state (240 h).
Figure 1B was unfixed biofilm scraped directly onto double-sided carbon tape, frozen while hydrated in liquid N2, splutter-coated with Platinum for 2 min, and imaged using a dual beam focused ion beam (FIB)-FE-SEM (Helios NanoLab, FEI Company, Hilsboro, OR, USA) equipped with a cryostage.
SAMPLE COLLECTION AND ANALYSIS
Reactor outflow was collected on ice and measured daily to monitor flow rate. Samples of the planktonic phase were collected at the outflow for optical density at 600 nm (OD), high performance liquid chromatography (HPLC), protein measurements, and direct cell counts. Filtered samples were analyzed in triplicate with a fucose internal standard, for lactate, acetate, and formate concentrations via HPLC (Agilent 1200 series) equipped with a BioRad Aminex HPX-87H column. Lactate and formate concentration were measured with a VWD detector while acetate concentration was measured with an RID detector. Planktonic cultures were centrifuged at 6,000 g for 10 min and the whole cell pellet was analyzed for protein, hexose, pentose, and uronic acid composition. Biofilm samples for these analyses were collected by aseptically replacing a biofilm coupon with a sterile butyl stopper, and scraping the biofilm into sterile water with a spatula. Whole biofilm was analyzed for protein and carbohydrates. Protein concentrations were determined with the Lowry et al. (1951) assay using bovine serum albumin as the standard. Hexose sugars were measured by the colorimetric cysteine–sulfuric acid method with glucose as the standard. Pentose sugars were measured with a colorimetric orcinol-FeCl3 assay with xylose as the standard. A colorimetric carbazole assay was used to measure uronic acid concentration with D-galacturonic acid as the standard (Chaplin and Kennedy, 1994).
RESULTS
BIOFILM STRUCTURE AND COMPOSITION
Monocultures of D. vulgaris formed biofilm on silica slides under continuous culture conditions when sulfate was provided as an electron acceptor (Figures 1A,C). Monoculture M. maripaludis did not form a biofilm on silica slides when grown in continuous culture supplemented with H2, as observed with protein assay, light microscopy, and scanning electron microscopy. Material was observed on the glass slides but was confirmed to be salts via energy dispersive X-ray spectroscopy and not protein or carbohydrate (data not shown).
When D. vulgaris and M. maripaludis were cocultivated with lactate (without sulfate and H2), methane was produced and biofilm was formed. The coculture biofilm had an altered appearance and structure compared to monoculture D. vulgaris biofilm (compare Figures 1A to 1B and 1C to 1D) and M. maripaludis cells were observed in both the biofilm and planktonic phases. These results indicate that the methanogen was dependent upon D. vulgaris under the tested conditions to grow in a biofilm state.
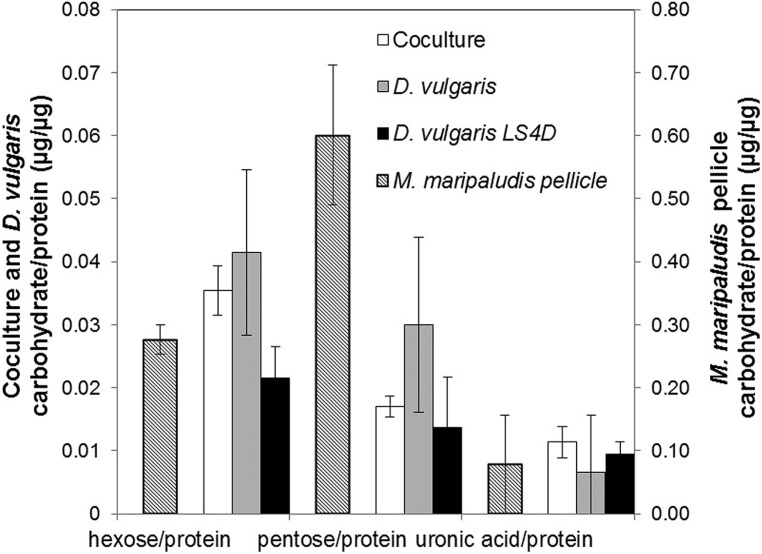
The protein and carbohydrate levels were compared for different growth conditions (cell-associated carbohydrate levels were normalized to protein biomass). As previously reported, D. vulgaris does not produce an extensive carbohydrate-rich biofilm on glass slides (Clark et al., 2007), but in CCM, D. vulgaris produced slightly increased levels of hexose and pentose equivalents compared to growth in LS4D medium (Figure 2). The uronic acid levels were similar for D. vulgaris when grown in LS4D or CCM (Figure 2) under the tested conditions. In the coculture biofilms, the uronic acid levels were similar while hexose and pentose levels were slightly decreased compared to D. vulgaris monocultures in CCM (Figure 2). The reported values were lower than previous reports for other monoculture and multispecies biofilm EPS that can constitute as much as 90% of the dry mass of a culture (Flemming and Wingender, 2010; Poli et al., 2010). Lack of extracellular material might present less mass transfer resistance to H2 diffusion and would therefore be beneficial to both organisms. As noted above, M. maripaludis did not form monoculture biofilm under the tested conditions; however, M. maripaludis did form a pellicle when grown in static tubes as a monoculture. The M. maripaludis pellicle had approximately 10-fold more uronic acid, 7-fold more hexose, and 30-fold more pentose compared to coculture biofilm (Figure 2). These results suggest that M. maripaludis had altered carbon flow that resulted in less carbon allocation to EPS.
FIGURE 2.
Biofilm carbohydrate composition normalized to protein biomass for hexose, pentose, and uronic acid in continuous culture biofilm (coculture and D. vulgaris monoculture, primary axis) and batch M. maripaludis pellicle (secondary axis). Error bars represent 95% confidence interval. Coculture in CCM n = 6, D. vulgaris in CCM n = 4, D. vulgaris in LS4D n = 8, M. maripaludis batch pellicle in CCM n = 3.
COCULTURE BIOFILM DEVELOPMENT AND STRUCTURE
Coculture biofilm was initiated by D. vulgaris, which formed a monolayer during the initial 48 h of batch mode in the continuous culture system (Figures 3A,D). At this early time point (0 h, initiation of flow) D. vulgaris out-numbered M. maripaludis in the biofilm 32:1, while the planktonic phase ratios of D. vulgaris to M. maripaludis were 2.7:1 (Table 1). After 48 h of continuous culture the biofilm grew in ridges, both normal and parallel to flow caused by liquid agitation (Figures 3B,E). Cell ratios in the biofilm decreased rapidly to 3.5:1 after 48 h as M. maripaludis cells were incorporated into the biofilm and grew, while planktonic ratios increased slightly to an average of 3.2:1. After 240 h, steady-state ratios of cells in the biofilm remained approximately 2.2:1 with similar planktonic ratios of 1.6:1 (Table 1). This is in contrast to the published 4:1 ratios observed in planktonic-only reactors for the same syntrophic pair under lactate-excess conditions and our own observation of 6.3:1 in planktonic phase-only reactors (Table 1; Stolyar et al., 2007; Walker et al., 2009, 2012). These results indicate that four times more D. vulgaris are typically needed to oxidize enough lactate to generate sufficient H2 for M. maripaludis in the bulk aqueous phase; however, in the presence of biofilm, a more even distribution (approximately 2:1) of interacting populations was sustained in both the biofilm and planktonic phases.
Table 1.
Percent of M. maripaludis and D. vulgaris cells in planktonic and biofilm phases over time [row 1 early (0 h), 2 intermediate (48 h), 3 steady-state (240 h)] in a reactor containing both growth phases, a reactor where biofilm has been removed (row 4), a reactor with an increased loading rate (row 5), and a reactor containing a coculture of ΔpilA mutant D. vulgaris with wild-type M. maripaludis (row 6).
| Rows | % M. maripaludis biofilm | % D. vulgaris biofilm | % M. maripaludis planktonic | % D. vulgaris planktonic | |
|---|---|---|---|---|---|
| 1 | Biofilm and planktonic early | 3% ± 0.8 | 97% ± 0.8 | 26% ± 13.7 | 69% ± 11.1 |
| 2 | Biofilm and planktonic intermediate | 22% ± 1.5 | 78% ± 1.5 | 23% ± 12.1 | 73% ± 10.6 |
| 3 | Biofilm and planktonic steady state | 31% ± 1.4 | 69% ±1.4 | 36% ±10.8 | 59% ±2.2 |
| 4 | WT planktonic only | 0 | 0 | 13% ± 1.9 | 82% ± 1.9 |
| 5 | Biofilm and planktonic increased loading rate | 18% ± 1.5 | 82% ± 1.5 | 22% ± 5.9 | 73% ± 3.3 |
| 6 | ΔpilA planktonic only | 0 | 0 | 19% ± 1.4 | 81% ± 1.4 |
Error is the 95% confidence interval. The number of images analyzed for each time point is early n = 146, intermediate n = 269, steady-state n = 2,118, biofilm removed n = 140, increased loading rate n = 174, and ΔpilA n = 120.
Coculture biofilm macrostructure was observed with both fixed and unfixed, still hydrated biofilm (Figures 1D and 3C,F). The structured biofilm included tall ridges and spires with deep valleys, often 300–400 μm tall, but always with at least one dimension <50 μm as measured by fluorescence microscopy of intact hydrated biofilm or cryosections of frozen hydrated biofilm (Figure 3F). Notably, the macrostructure was not observed in D. vulgaris monoculture biofilms grown in LS4D medium (Clark et al., 2007) nor in CCM (Figures 1A,C). The critical biofilm thickness that would allow for diffusion of 30 mM lactate to the substratum was estimated to be 50 μm, as predicted by a 1D BAM (Figure S3 and Table S1). These results suggest that the macrostructure was influenced by lactate diffusion limitation. Microcolonies of M. maripaludis cells were spread throughout a matrix of D. vulgaris cells with an intermixed pattern. Cell association in the structured biofilm was observed to be random with no pattern of colocalization detectable (Figure S4). It has recently been shown that increased intermixing is a marker of cooperation (Momeni et al., 2013), so it is reasonable that this cooperative community was highly intermixed.
BIOFILM AND PLANKTONIC COMMUNITY FUNCTION: THE BASE CASE
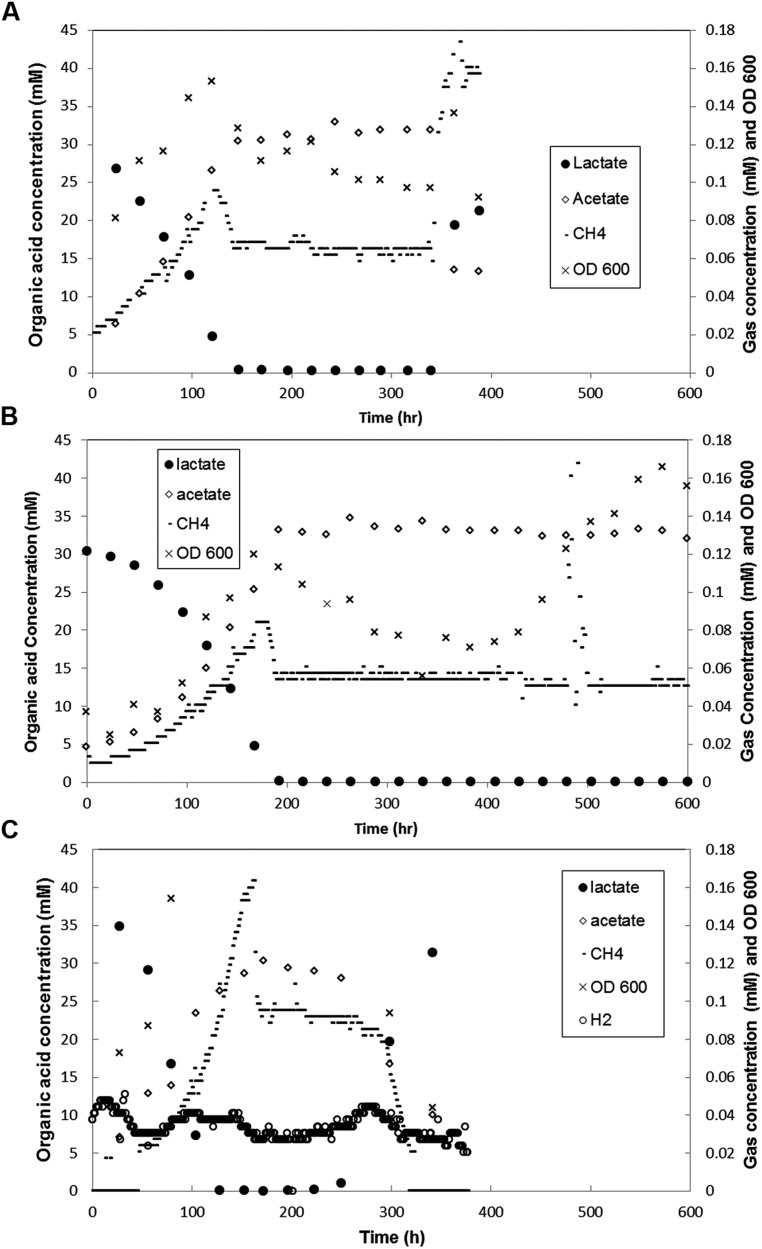
The base case represents the standard syntrophic system described above that contained a structured biofilm and a planktonic phase in continuous culture. Little, if any previous work has been done to characterize syntrophic interactions with interacting biofilm and planktonic phases, so a baseline understanding of function (CH4 and H2 production and lactate consumption) was necessary to determine a basal state under the tested conditions. During the first 100 h of biofilm development, methane levels increased as lactate levels declined with an equimolar increase in acetate (Figure 4A). The biofilm population was 78% D. vulgaris and 22% M. maripaludis (3.5:1) with similar planktonic population distribution (Table 1). During the next 50 h, the system approached a steady-state in which all 30 mM of lactate was consumed, and both organisms in both phases of the reactor increased rapidly in number. As lactate became limiting, the OD and methane concentration peaked for one retention time of the reactor, and then declined to a steady-state. The methane concentration stabilized but OD continually decreased while biofilm biomass increased for another 100 h after lactate was not detectable. These results indicate that biofilm cells were competitive for bulk-phase lactate, and that the biofilm growth mode contributed to more efficient, multi-species substrate utilization. This is further demonstrated in that H2 was not detectable in the reactor headspace at any point. Walker et al. (2009) observed a spike (50 Pa) followed by a constant low level of H2 (less than 10 Pa) at steady-state when a planktonic-only system was not limited for lactate. Presumably all H2 produced in our biofilm reactor was efficiently consumed. It should also be noted that H2 was not detected when the lactate loading rate was increased (discussed below).
FIGURE 4.
Metabolite concentration over time in coculture biofilm reactors (A) with both biofilm and planktonic phases in which loading rate was increased at 341 h, (B) that had biofilm coupons removed at 432 h (C) grown with ΔpilA mutant D. vulgaris and wild-type M. maripaludis. Each graph is representative of one of two duplicate experiments.
In aerobic biofilms, it has been shown that cells near the substratum can be limited in oxygen and metabolically inactive, i.e., not all the biofilm biomass is active (Xu et al., 2000). To assess the metabolic state of the syntrophic biofilm, intact, steady-state coculture biofilm was incubated with CTC (Stewart et al., 1994). The validity of this method for anaerobes has been debated with the primary concern that CTC is abiotically reduced in the presence of sulfide and cysteine (Stewart et al., 1994; Gruden et al., 2003; Halan et al., 2012). CT-formazan granules formed abiotically are poorly localized and rapidly photo-bleach, while CT-formazan of biogenic origin is an intracellular granule that is more resistant to photo-bleaching (Gruden et al., 2003). CCM contains 1 mM each of sodium sulfide and cysteine, but the biofilm was rinsed anoxically prior to staining in an anaerobic glovebag. The incubated biofilm was directly observed with CSLM, and the biofilm biomass showed respiratory potential based on formation of CT-formazan. However, portions of the intact biofilm were not visible via CSLM due to depth limitations (Figure 5A), therefore the biofilm was scraped for visualization (post-staining). Upon inspection of scraped biofilm, the entire biofilm biomass was stained (Figures 5B,C), and reduced CT-formazan granules could be observed in nearly all cells, localized inside the cell with persistent fluorescence. These results indicate that the entire steady-state biofilm biomass retained respiratory potential and was metabolically active. The same results were obtained using FISH, where all biofilm cells exhibited strong fluorescence irrespective of location, and these results corroborated the idea that all cells were active (Figure 5D).
FIGURE 5.
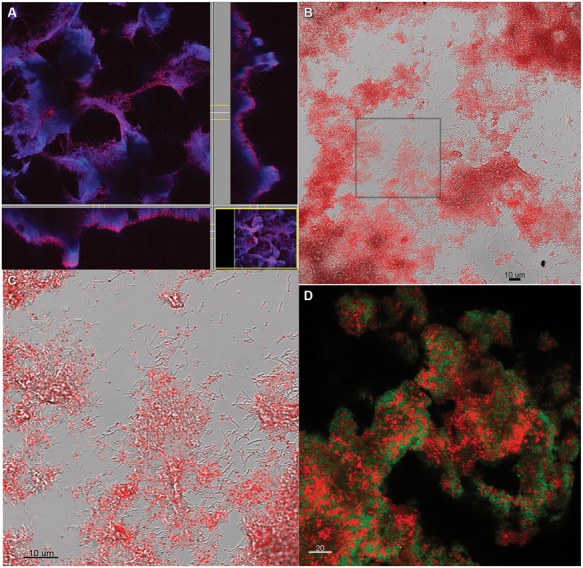
Coculture biofilm (A) stained with CTC while intact and hydrated and showing all biomass stained with DAPI in blue, and CTC in red, or purple where both DAPI and CTC are present (B) scraped from the slide after CTC staining. (C) Zoomed in from the inset in (B) showing individual grains of red fluorescent CT-formazan in each cell. (D) Coculture biofilm scraped from the substratum, fixed and hybridized with domain-specific probes for D. vulgaris (green) and M. maripaludis (red).
INCREASED LOADING RATE
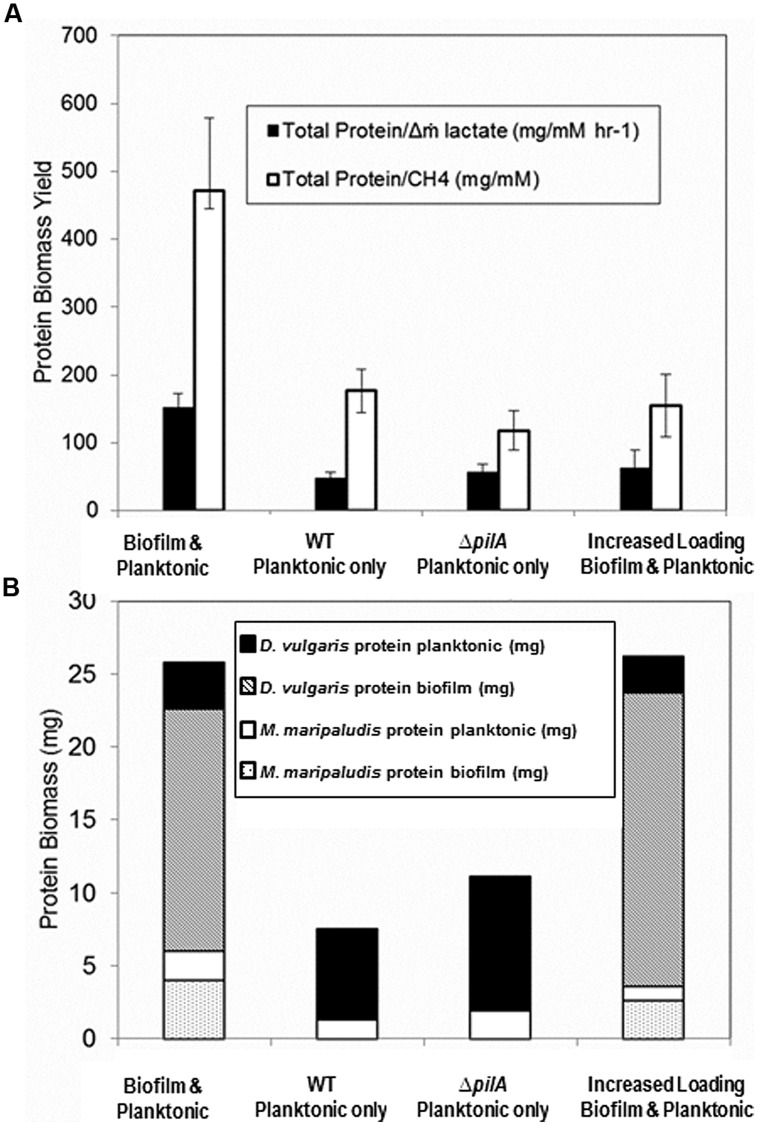
To test the effect of a sudden input of nutrients on a stable community, the dilution rate was increased after 341 h to 0.109 h-1 (approximately 6-fold) in a biofilm reactor in steady-state. Methane levels increased within 1 h and peaked at ∼0.17 mM after 27 h (Figure 4A). The optical density increased slightly and then declined to just below steady-state levels, with an ∼20% decrease of D. vulgaris cells and ∼15% decrease in M. maripaludis cells in the planktonic phase based on cell counts. The system was monitored for four retention times (RT = 9.9 h) with biofilm samples removed after 48 h, and the decreased planktonic biomass is likely a result of washout. The doubling time of D. vulgaris in CCM supplemented with sulfate is approximately 20 h (k = 0.04 h-1) while the doubling time of M. maripaludis with unlimited H2 in CCM is 5 h (k = 0.14 h-1). Under these conditions as D. vulgaris was washed out of the reactor, H2 was not produced at a rate that would allow the methanogen to divide before the entire reactor was turned over in 9.9 h. The total amount of biomass in the reactor did not change with increased loading, and while the planktonic populations began to washout, biomass was balanced by growth in the biofilm (Figure 6B). Under these conditions, biomass distribution in the biofilm shifted toward a greater percentage of D. vulgaris that increased by nearly 25% from 16.5 to 20.0 mg (Figure 6B), and this resembled pre-steady-state (i.e., when lactate was not entirely consumed) population structure most likely as a result of the increased loading rate for lactate (∼3 mM h-1; Table 1). Despite the altered population ratio, the macrostructure of the biofilm remained similar to the steady-state structure (lactate loading rate ∼0.5 mM h-1) with a D. vulgaris matrix intermixed with M. maripaludis.
FIGURE 6.
(A) Biomass yield per lactate mass flux and per methane for reactors containing coculture with biofilm and planktonic phases (base case; n = 8), planktonic phase only after biofilm was removed (n = 14), ΔpilA mutant coculture planktonic phase only (n = 7), and both biofilm and planktonic phases at an increased loading rate (n = 2). Error bars represent 95% confidence interval. (B) Biomass distribution of each cell type in biofilm and planktonic phases for the same culture conditions based upon cell volume ratios and cell counts respectively. Unpaired two-tailed t-test results indicate total protein/Δm lactate difference is significant between coculture biofilm and planktonic (base case) versus all other conditions (p < 0.005), but that planktonic only versus increased loading rate is not significant (p = 0.4). Total Protein/CH4 unpaired two-tailed t-test results were significant between biofilm and planktonic (base case) versus all other conditions [versus WT planktonic and ΔpilA planktonic (p < 0.005), biofilm and planktonic (base case) versus increased loading rate (p = 0.05), planktonic only versus increased loading rate (p = 0.4)].
In spite of lactate-excess, H2 was still undetectable (limit of detection ∼0.0001 mM or 0.25 Pa) when the loading rate was increased. This is contrary to published results for excess lactate planktonic-only conditions where H2 was continually 5–10 Pa at steady-state (Walker et al., 2009). These results suggest that the presence of biofilm caused a more efficient consumption or transfer of the produced H2 gas between D. vulgaris and M. maripaludis. Biomass yield per methane produced (mg protein/mM CH4) was significantly lower in lactate-excess conditions than in lactate-limited conditions (Figure 6A) and the same was true for biomass yield per lactate mass flux (mg protein/mM h-1 lactate). Essentially the same amount of biomass or carrying capacity was actively maintained under both conditions (i.e., lactate-excess versus lactate-limited) but the population distributions were different, and biofilm was able to increase metabolic flux without an increase in biomass. The carrying capacity, K, is defined as the maximum potential population size a given landscape is capable of supporting and is a common attribute used to describe population dynamics in ecology (Stilling, 2003; Berck et al., 2012). When additional lactate was available via an increased loading rate, the system was perturbed (washout of planktonic biomass) but the total biofilm biomass increased (Figure 6B). The total carrying capacity thus remained the same even though the population distribution and metabolic flux changed under lactate-excess conditions.
BIOFILM REMOVAL
In a separate biofilm reactor at steady-state, the biofilm coupons were removed after 432 h (Figure 4B) to test the stability and population structure of the planktonic community in the absence of biofilm. Upon biofilm removal, the planktonic optical density increased within 24 h, but methane levels did not increase for 50 h (Figure 4B). After a 50-h static period, methane concentrations increased rapidly but declined back to original steady-state levels (15 h time period). Lactate and H2 were not detected, the OD increased, and similar levels of methane were produced as the system attempted to reach a new steady-state. The planktonic-phase only reactor population was 82% D. vulgaris but both D. vulgaris and M. maripaludis increased in absolute number based on cell counts and OD (Table 1 and Figure 4B). Although 1.4 times greater biomass was maintained in the planktonic phase alone than the planktonic phase of the base case, the total reactor biomass was 3.4 times lower than the total biofilm plus planktonic biomass in the base case reactor (Figure 6B). The biomass yield was significantly lower than in the base case, and similar to total biomass yields in lactate-excess conditions (Figure 6A). Without a biofilm community, the carrying capacity of the system was significantly reduced under lactate-limiting conditions and the population distribution was less even.
To further investigate the role of biofilm in carrying capacity and stability, a mutant D. vulgaris, ΔpilA, was grown in coculture with wild-type M. maripaludis. The ΔpilA D. vulgaris lacks a presumptive type IV pilus and is deficient in biofilm formation (Figure S5). In batch coculture experiments, total methane production and growth were the same as wild-type coculture (data not shown). However, in continuous culture, the ΔpilA coculture did not form biofilm and the coculture grew as a planktonic-phase with similar levels of methane produced compared to wild-type coculture (0.09 mM at steady-state; Figure 4C). At 175 h, the biomass yield of ΔpilA coculture was similar to the planktonic-only wild-type coculture reactor (Figure 6A), and the total biomass was slightly higher than planktonic-only wild-type coculture (Figure 6B) with a similar distribution of D. vulgaris (Table 1). However, the mutant coculture did not stabilize and completely washed out of the reactor within 400 h (Figure 4C). In addition, in contrast to wild-type, H2 was detectable (50–120 Pa) over the whole 350 h of continuous culture until cells were too few to count in the planktonic phase (Figure 4C). This result further demonstrates the role of biofilm structure in facilitating and stabilizing syntrophic interactions. In the absence of biofilm (either physically removing the biofilm or biofilm deficient coculture), the community was not stable, H2 production and consumption were not balanced, and the carrying capacity declined for the methanogen.
In another δ-Proteobacterium, Geobacter sulfurreducens, PilA has been shown to be involved in extracellular electron transfer and biofilm formation (Richter et al., 2012), while in an aerobic δ-Proteobacterium, Myxococcus xanthus, PilA was shown to interact with biofilm EPS (Wei et al., 2012). Further work is needed to determine the role of type IV pili in facilitating interactions between SRB and hydrogenotrophic methanogens, but it seems likely that the pilus functions in attachment of cells to surfaces (biotic or abiotic) that directly or indirectly facilitates metabolic exchange.
DISCUSSION
The primary objective of this work was to characterize the relationship between function and structure of a syntrophic biofilm community. Previous work suggests that specific structural patterns can be expected in interacting communities (Nielsen et al., 2000; Gu et al., 2013; Momeni et al., 2013), and that these patterns are dependent upon the nature of the interaction. We observed a structured syntrophic biofilm with complex ridges and channels, where both partners were highly intermixed. Similar biofilm structures have been observed in mixed communities and are also presumed to be a direct result of interaction type (Nielsen et al., 2000; Molin and Tolker-Nielsen, 2003), so it is reasonable to expect that structure affects community function and vice versa. Several results reported here indicate that syntrophic lactate oxidation and transfer of the H2 intermediate dictated the biofilm structure. Biofilm structures were never observed to exceed 50 μm in at least one dimension, and the BAM for this community predicted that biofilm thicker than 50 μm would experience lactate diffusion limitation at the substratum. These results suggest that lactate diffusion governed D. vulgaris biofilm structure. This type of structure would also have the same positive effect on H2 diffusing away from the SRB, where a buildup of the inhibitory by-product would prevent further lactate oxidation. Monoculture biofilms of D. vulgaris grown with sulfate and lactate similarly form only thin biofilms, yet they do not form tall structures in the way that syntrophic D. vulgaris does. These results suggest that syntrophic interactions drive the observed structural features.
When we consider the structure–function relationship from the point of the methanogen, the results suggest that the biofilm structure was driven by H2. In monoculture with H2, M. maripaludis did not form biofilm, however, when grown syntrophically the methanogen did join the H2-producing SRB biofilm. While it is possible that D. vulgaris biofilm simply provided a more suitable surface for attachment of M. maripaludis, it is also very likely that H2 drove the interaction specifically. We have recently shown chemotaxis toward H2 gas (hydrogenotaxis) in M. maripaludis (Brileya et al., 2013) and the response is especially strong under H2-limited conditions. H2 produced by SRB in the biofilm could diffuse to the aqueous and gas phases of the reactor, making it possible for planktonic M. maripaludis to scavenge the energy source without joining the biofilm. In spite of this option, more methanogen biomass was observed in the biofilm than the planktonic phase, whenever biofilm was present in the system. This suggests that some benefit is gained by interacting directly or closely with the SRB in the biofilm. These observations are supported by the lack of H2 detected during cultivation of wild-type populations as coculture biofilm. The results of the mutant coculture experiment further support a benefit from close interaction, since the mutant SRB lacked a pilus that presumably helped interactions directly through attachment or motility in the biofilm. The lack of direct interaction in biofilm resulted in detectable H2, and therefore inefficient transfer of the intermediate. It was recently shown that motility is an important determinant for structuring mixed biofilms when a motile Bacillus could infiltrate a Staphylococcus biofilm (Houry et al., 2012).
Analysis of cell-associated carbohydrates showed that M. maripaludis likely reallocates carbon in the syntrophic biofilm, given that the coculture biofilm had 10-fold less cell-associated carbohydrate than a monoculture M. maripaludis pellicle. One possible explanation is that a thick extracellular matrix would increase H2 mass transfer resistance, so the methanogen produced less carbohydrate to facilitate in H2 diffusion through the biofilm. Another possible explanation for altered carbon allocation is that M. maripaludis produced less EPS when grown as syntrophic biofilm to facilitate repositioning within the biofilm. M. maripaludis has only been shown to sense and swim toward H2 gas in liquid (Brileya et al., 2013), but it has not been shown to swarm, so it remains unclear whether individuals could move through a dense EPS matrix toward a higher concentration of H2.
Pure mutualism refers to the fact that the relationship is obligate and the growth rates of both populations are limited only by the concentrations of critical substrates produced by the partner (Meyer et al., 1975). The case of a SRB that oxidizes organic carbon to H2 and a methanogen that consumes the H2 is a variation that can be termed mutualism via product inhibition (Dean, 1985). Historically, microbial interactions have been studied in terms of competition for substrate and the competition coefficients are typically a ratio of the yield coefficients (Dean, 1985). Thus, stable or even unstable equilibria do not exist in terms of one population ‘winning’ over the other. However, these equations are based upon chemostats with only bulk-phase populations and not biofilms with inherent variability. Positive interactions could stabilize many more microbial interactions than previously thought (Shindala et al., 1965; Megee et al., 1972), particularly for biofilms. Our results support that community stability is a result of syntrophic interaction in biofilm. When lactate-loading rate was increased, causing washout conditions in the aqueous phase, biomass in the biofilm increased while planktonic biomass decreased. The washout situation highlights a complication of mutualistic interaction under flow conditions in which a population with a slower specific growth rate produces the limiting substrate of another population. Biomass retention in biofilm and close interaction represent logical ecological solutions to this problem. Community members are able to stay in a desirable location, rather than be washed away to potentially unfavorable environments. The mutant coculture could not form biofilm, and possibly close interactions, and therefore was unable to form a stable syntrophy with tight coupling between H2 production and consumption.
Macroecologists and microbial ecologists alike have modeled mutualism to gain insight into inter-population dynamics, and results predict stability of cooperative populations under only specific density-dependent conditions (Boucher, 1988). Experiments with mutualistic microorganisms have revealed many adaptations to syntrophic relationships, including alternative electron-transport pathways, differences in gene expression patterns in the presence of a syntrophic partner, and rapid evolution resulting in optimized biomass production (Shimoyama et al., 2009; Walker et al., 2009, 2012; Hillesland and Stahl, 2010; Plugge et al., 2011; Bernstein et al., 2012; Lawrence et al., 2012; Sieber et al., 2012). In this syntrophic system, while we observed that biofilm growth mode promoted the greatest biomass retention and allowed the system to reach increased carrying capacity, we also observed that this biomass was metabolically functional, in spite of lactate limitation. CTC staining and FISH indicated that the whole biofilm biomass had respiratory potential. When additional lactate was added via an increased loading rate, the biofilm community responded within 1 h by increasing electron flux from lactate to methane. It is quite interesting to consider this result in the context of a low-nutrient environment, where it seems likely that a natural biofilm community could remain poised for episodic nutrient availability. Our results indicate that in a mixed community, syntrophs are able to rapidly cycle electrons or carbon.
In this model syntrophic system, structured biofilm promoted maximum carrying capacity, contributed to cooperative resource sharing (i.e., improved H2 transfer) and provided greater community stability when compared to planktonic-only populations. Although both biofilms and syntrophic communities are inherently variable and heterogeneous, these culture conditions are environmentally relevant. Mixed culture biofilm reactors can be used to experimentally explore ecological and evolutionary phenomena in a more constrained setting. It remains to be seen what genetic and metabolic controls are responsible for the observed responses in this system, and future work is planned to understand how specific biofilm structures and interactions can impact meso- and macro-scale processes including greenhouse gas production, biogeochemical cycling, and waste conversion.
AUTHOR CONTRIBUTIONS
Kristen A. Brileya designed and performed experiments, analyzed and interpreted data, drafted and revised the manuscript. Laura B. Camilleri performed experiments, analyzed and interpreted data, and revised the manuscript. Grant M. Zane designed and performed experiments to create the mutant ΔpilA D. vulgaris, and revised the manuscript. Judy D. Wall designed experiments to create the mutant ΔpilA D. vulgaris, interpreted the data for this manuscript, and made critical revisions to the manuscript. Matthew W. Fields designed experiments, interpreted data, drafted and revised the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are grateful for thoughtful discussions with Dr. Chris Walker, Dr. Sergey Stolyar, and Prof. David Stahl. Thanks to the Stahl laboratory for cultures of M. maripaludis. We are deeply indebted to Betsey Pitts for tirelessly sharing her knowledge of confocal and fluorescence microscopy and image analysis, and also to Dr. Roland Hatzenpichler and Dr. Sebastian Lücker for their suggestions and discussions about FISH and 3D-FISH. We thank Dr. Ross Carlson for the use of HPLC with much appreciated assistance from Dr. Hans Bernstein and Kris Hunt. This work was supported as a component of ENIGMA, a scientific focus area program supported by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research, Genomics: GTL Foundational Science through contract DE-AC02-05CH11231 between Lawrence Berkeley National Laboratory and the U.S. Department of Energy. Kristen A. Brileya and Laura B. Camilleri were additionally supported by a NSF-IGERT fellowship in Geobiological Systems at Montana State University (DGE 0654336). The confocal microscope equipment used was purchased with funding from the NSF-Major Research Instrumentation Program and the M.J. Murdoch Charitable Trust. The FIB-SEM image was acquired with the help of Alice Dohnalkova and Bruce Arey through a user proposal funded by the Environmental Molecular Sciences Laboratory at Pacific Northwest National Lab. Zeiss FE-SEM images were acquired using equipment in the Image and Chemical Analysis Laboratory (ICAL) at Montana State University.
SUPPLEMENTARY MATERIAL
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fmicb.2014.00693/abstract
REFERENCES
- Al-Ahmad A., Follo M., Selzer A. C., Hellwig E., Hannig M., Hannig C. (2009). Bacterial colonization of enamel in situ investigated using fluorescence in situ hybridization. J. Med. Microbiol. 58 1359–1366 10.1099/jmm.0.011213-0 [DOI] [PubMed] [Google Scholar]
- Berck P., Levy A., Chowdhury K. (2012). An analysis of the world’s environment and population dynamics with varying carrying capacity, concerns and skepticism. Ecol. Econ. 73 103–112 10.1016/j.ecolecon.2011.09.019 [DOI] [Google Scholar]
- Bernstein H. C., Paulson S. D., Carlson R. P. (2012). Synthetic Escherichia coli consortia engineered for syntrophy demonstrate enhanced biomass productivity. J. Biotech. 157 159–166 10.1016/j.jbiotec.2011.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher D. H. (1988). The Biology of Mutualism. New York: Oxford University Press. [Google Scholar]
- Brenner K., Arnold F. H. (2011). Self-organization, layered structure, and aggregation enhance persistence of a synthetic biofilm consortium. PLoS ONE 6:e16791 10.1371/journal.pone.0016791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brileya K. A., Camilleri L. B., Fields M. W. (2014). “3D-Fluorescence in situ hybridization of intact, anaerobic biofilm,” in Engineering and Analyzing Multicellular Systems, eds Sun L., Shou W. (New York: Springer; ), 189–197. [DOI] [PubMed] [Google Scholar]
- Brileya K. A., Connolly J. M., Downey C., Gerlach R., Fields M. W. (2013). Chemotaxis toward hydrogen gas by Methanococcus maripaludis. Sci. Rep. 3 1–7 10.1038/srep03140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant M., Campbell L., Reddy C., Crabill M. (1977). Growth of Desulfovibrio in lactate or ethanol media low in sulfate in association with H2-utilizing methanogenic bacteria. Appl. Environ. Microbiol. 33 1162–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canfield D. E., Stewart F. J., Thamdrup B., De Brabandere L., Dalsgaard T., Delong E. F., et al. (2010). A cryptic sulfur cycle in oxygen-minimum-zone waters off the Chilean coast. Science 330 1375–1378 10.1126/science.1196889 [DOI] [PubMed] [Google Scholar]
- Chaplin M. F., Kennedy J. F. (1994). Carbohydrate Analysis: A Practical Approach, 2nd Edn Oxford: Irl Press. [Google Scholar]
- Clark M. E., Edelmann R. E., Duley M. L., Wall J. D., Fields M. W. (2007). Biofilm formation in Desulfovibrio vulgaris Hildenborough is dependent upon protein filaments. Environ. Microbiol. 9 2844–2854 10.1111/j.1462-2920.2007.01398.x [DOI] [PubMed] [Google Scholar]
- Clark M. E., He Q., He Z., Huang K. H., Alm E. J., Wan X. F., et al. (2006). Temporal transcriptomic analysis as Desulfovibrio vulgaris Hildenborough transitions into stationary phase during electron donor depletion. Appl. Environ. Microbiol. 72 5578–5588 10.1128/AEM.00284-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark M. E., He Z., Redding A. M., Joachimiak M. P., Keasling J. D., Zhou J. Z., et al. (2012). Transcriptomic and proteomic analyses of Desulfovibrio vulgaris biofilms: carbon and energy flow contribute to the distinct biofilm growth state. BMC Genomics 13:138 10.1186/1471-2164-13-138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daims H., Lücker S., Wagner M. (2006). daime, a novel image analysis program for microbial ecology and biofilm research. Environ. Microbiol. 8 200–213 10.1111/j.1462-2920.2005.00880.x [DOI] [PubMed] [Google Scholar]
- Dean A. M. (1985). “The dynamics of microbial commensalisms and mutualisms,” in The Biology of Mutualism ed. Boucher D. H. (Oxford: Oxford University Press; ), 270–300. [Google Scholar]
- Flemming H.-C., Wingender J. (2010). The biofilm matrix. Nat. Rev. Microbiol. 8 623–633. [DOI] [PubMed] [Google Scholar]
- Gokhale C. S., Traulsen A. (2012). Mutualism and evolutionary multiplayer games: revisiting the Red King. Proc. Biol. Sci. 279 4611–4616 10.1098/rspb.2012.1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross R., Hauer B., Otto K., Schmid A. (2007). Microbial biofilms: new catalysts for maximizing productivity of long-term biotransformations. Biotechnol. Bioeng. 98 1123–1134 10.1002/bit.21547 [DOI] [PubMed] [Google Scholar]
- Gruden C., Fevig S., Abu-Dalo M. (2003). 5-Cyano-2, 3-ditolyl tetrazolium chloride (CTC) reduction in a mesophilic anaerobic digester: measuring redox behavior, differentiating abiotic reduction, and comparing FISH response as an activity indicator. J. Microbiol. Methods 52 59–68 10.1016/S0167-7012(02)00134-3 [DOI] [PubMed] [Google Scholar]
- Gu H., Hou S., Yongyat C., De Tore S., Ren D. (2013). Patterned biofilm formation reveals a mechanism for structural heterogeneity in bacterial biofilms. Langmuir 29 11145–11153 10.1021/la402608z [DOI] [PubMed] [Google Scholar]
- Halan B., Buehler K., Schmid A. (2012). Biofilms as living catalysts in continuous chemical syntheses. Trends Biotech. 30 453–465 10.1016/j.tibtech.2012.05.003 [DOI] [PubMed] [Google Scholar]
- Hall-Stoodley L., Costerton J., Stoodley P. (2004). Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2 95–108 10.1038/nrmicro821 [DOI] [PubMed] [Google Scholar]
- Hall-Stoodley L., Hu F. Z., Gieseke A., Nistico L., Nguyen D., Hayes J., et al. (2006). Direct detection of bacterial biofilms on the middle-ear mucosa of children with chronic otitis media. JAMA 296 202–211 10.1001/jama.296.2.202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen S. K., Rainey P. B., Haagensen J. A. J., Molin S. (2007). Evolution of species interactions in a biofilm community. Nature 445 533–536 10.1038/nature05514 [DOI] [PubMed] [Google Scholar]
- Hillesland K. L., Stahl D. A. (2010). Rapid evolution of stability and productivity at the origin of a microbial mutualism. Proc. Natl. Acad. Sci. U.S.A. 107 2124–2129 10.1073/pnas.0908456107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houry A., Gohar M., Deschamps J., Tischenko E., Aymerich S., Gruss A., et al. (2012). Bacterial swimmers that infiltrate and take over the biofilm matrix. Proc. Natl. Acad. Sci. U.S.A. 109 13088–13093 10.1073/pnas.1200791109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubovics N. S. (2010). Talk of the town: interspecies communication in oral biofilms. Mol. Oral Microbiol. 25 4–14 10.1111/j.2041-1014.2009.00563.x [DOI] [PubMed] [Google Scholar]
- Kato S., Hashimoto K., Watanabe K. (2011). Methanogenesis facilitated by electric syntrophy via (semi)conductive iron-oxide minerals. Environ. Microbiol. 14 1646–1654 10.1111/j.1462-2920.2011.02611.x [DOI] [PubMed] [Google Scholar]
- Kato S., Watanabe K. (2010). Ecological and evolutionary interactions in syntrophic methanogenic consortia. Microbes Environ. 25 145–151 10.1264/jsme2.ME10122 [DOI] [PubMed] [Google Scholar]
- Lawrence D., Fiegna F., Behrends V., Bundy J. G., Phillimore A. B., Bell T., et al. (2012). Species interactions alter evolutionary responses to a novel environment. PLoS Biol. 10:e1001330 10.1371/journal.pbio.1001330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leloup J., Fossing H., Kohls K., Holmkvist L., Borowski C., Jørgensen B. B. (2009). Sulfate-reducing bacteria in marine sediment (Aarhus Bay, Denmark): abundance and diversity related to geochemical zonation. Environ. Microbiol. 11 1278–1291 10.1111/j.1462-2920.2008.01855.x [DOI] [PubMed] [Google Scholar]
- Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J. (1951). Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193 265–275. [PubMed] [Google Scholar]
- McInerney M. J., Sieber J. R., Gunsalus R. P. (2009). Syntrophy in anaerobic global carbon cycles. Curr. Opin. Biotechnol. 20 623–632 10.1016/j.copbio.2009.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megee R. D., III, Drake J. F., Fredrickson A. G., Tsushiya H. M. (1972). Studies in intermicrobial symbiosis, Saccharomyces cerevisiae and Lactobacillus casei. Can. J. Microbiol. 18 1733–1742 10.1139/m72-269 [DOI] [PubMed] [Google Scholar]
- Meyer J. S., Tsuchiya H. M., Fredrickson A. G. (1975). Dynamics of mixed populations having complementary metabolism. Biotechnol. Bioeng. 17 1065–1081 10.1002/bit.260170709 [DOI] [Google Scholar]
- Moissl-Eichinger C., Huber H. (2011). Archaeal symbionts and parasites. Curr. Opin. Microbiol. 14 364–370 10.1016/j.mib.2011.04.016 [DOI] [PubMed] [Google Scholar]
- Molin S., Tolker-Nielsen T. (2003). Gene transfer occurs with enhanced efficiency in biofilms and induces enhanced stabilisation of the biofilm structure. Curr. Opin. Biotech. 14 255–261 10.1016/S0958-1669(03)00036-3 [DOI] [PubMed] [Google Scholar]
- Møller S., Sternberg C., Andersen J. B., Christensen B. B., Ramos J. L., Givskov M., et al. (1998). In situ gene expression in mixed-culture biofilms: evidence of metabolic interactions between community members. App. Environ. Microbiol. 64 721–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momeni B., Brileya K. A., Fields M. W., Shou W. (2013). Strong inter-population cooperation leads to partner intermixing in microbial communities. Elife 2:e00230 10.7554/eLife.00230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau J. W., Zierenberg R. A., Banfield J. F. (2010). Diversity of dissimilatory sulfite reductase genes (dsrAB) in a salt marsh impacted by long-term acid mine drainage. Appl. Environ. Microbiol. 76 4819–4828 10.1128/AEM.03006-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neef L., van Weele M., van Velthoven P. (2010). Optimal estimation of the present-day global methane budget. Global Biogeochem. Cycles 24 1–10 10.1029/2009GB003661 [DOI] [Google Scholar]
- Nielsen A. T., Tolker-Nielsen T., Barken K. B., Molin S. (2000). Role of commensal relationships on the spatial structure of a surface-attached microbial consortium. Environ. Microbiol. 2 59–68 10.1046/j.1462-2920.2000.00084.x [DOI] [PubMed] [Google Scholar]
- Perez-Osorio A. C., Williamson K. S., Franklin M. J. (2010). Heterogeneous rpoS and rhlR mRNA levels and 16S rRNA/rDNA (rRNA gene) ratios within Pseudomonas aeruginosa biofilms, sampled by laser capture microdissection. J. Bacteriol. 192 2991–3000 10.1128/JB.01598-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plugge C. M., Zhang W., Scholten J. C. M., Stams A. J. M. (2011). Metabolic flexibility of sulfate-reducing bacteria. Front. Microbiol. 2:81 10.3389/fmicb.2011.00081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poli A., Anzelmo G., Nicolaus B. (2010). Bacterial exopolysaccharides from extreme marine habitats: production, characterization and biological activities. Mar. Drugs 8 1779–1802 10.3390/md8061779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purdy K. J., Embley T. M., Nedwell D. B. (2002). The distribution and activity of sulphate reducing bacteria in estuarine and coastal marine sediments. Antonie Van Leeuwenhoek 81 181–187 10.1023/A:1020550215012 [DOI] [PubMed] [Google Scholar]
- Raskin L., Rittmann B. E., Stahl D. A. (1996). Competition and coexistence of sulfate-reducing and methanogenic populations in anaerobic biofilms. Appl. Environ. Microbiol. 62 3847–3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter L. V., Sandler S. J., Weis R. M. (2012). Two isoforms of Geobacter sulfurreducens PilA have distinct roles in pilus biogenesis, cytochrome localization, extracellular electron transfer, and biofilm formation. J. Bacteriol. 194 2551–2563 10.1128/JB.06366-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesinger W. H., Bernhardt E. S. (2013). Biogeochemistry: An Analysis of Global Change, 3rd Edn. Waltham, MA: Academic Press. [Google Scholar]
- Shimoyama T., Kato S., Ishii S., Watanabe K. (2009). Flagellum mediates symbiosis. Science 323 1574 10.1126/science.1170086 [DOI] [PubMed] [Google Scholar]
- Shindala A., Bungay H. R., III, Krieg N. R., Culbert K. (1965). Mixed-culture interactions I. Commensalism of Proteus vulgaris with Saccharomyces cerevisiae in continuous culture. J. Bacteriol. 89 693–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber J. R., McInerney M. J., Gunsalus R. P. (2012). Genomic insights into syntrophy: the paradigm for anaerobic metabolic cooperation. Annu. Rev. Microbiol. 66 429–452 10.1146/annurev-micro-090110-102844 [DOI] [PubMed] [Google Scholar]
- Stams A. J. M., Plugge C. M. (2009). Electron transfer in syntrophic communities of anaerobic bacteria and archaea. Nat. Rev. Microbiol. 7 568–577 10.1038/nrmicro2166 [DOI] [PubMed] [Google Scholar]
- Stewart P. S. (2003). Diffusion in biofilms. J. Bacteriol. 185 1485–1491 10.1128/JB.185.5.1485-1491.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart P. S., Franklin M. J. (2008). Physiological heterogeneity in biofilms. Nat. Rev. Microbiol. 6 199–210 10.1038/nrmicro1838 [DOI] [PubMed] [Google Scholar]
- Stewart P. S., Griebe T., Srinivasan R., Chen C. I., Yu F. P., deBeer D., et al. (1994). Comparison of respiratory activity and culturability during monochloramine disinfection of binary population biofilms. Appl. Environ. Microbiol. 60 1690–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stilling P. D. (2003). Ecology, 4th Edn. Upper Saddle River, NJ: Prentice Hall Inc. [Google Scholar]
- Stoecker K., Dorninger C., Daims H., Wagner M. (2010). Double labeling of oligonucleotide probes for fluorescence in situ hybridization (DOPE-FISH) improves signal intensity and increases rRNA accessibility. Appl. Environ. Microbiol. 76 922–926 10.1128/AEM.02456-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolyar S., Van Dien S., Hillesland K. L., Pinel N., Lie T. J., Leigh J. A., et al. (2007). Metabolic modeling of a mutualistic microbial community. Mol. Syst. Biol. 3 1–14 10.1038/msb4100131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thauer R. K., Kaster A.-K., Seedorf H., Buckel W., Hedderich R. (2008). Methanogenic archaea: ecologically relevant differences in energy conservation. Nat. Rev. Microbiol. 6 579–591 10.1038/nrmicro1931 [DOI] [PubMed] [Google Scholar]
- van Groenigen K. J., Osenberg C. W., Hungate B. A. (2012). Increased soil emissions of potent greenhouse gases under increased atmospheric CO2. Nature 475 214–216 10.1038/nature10176 [DOI] [PubMed] [Google Scholar]
- Walker C. B., He Z., Yang Z. K., Ringbauer J. A., He Q., Zhou J., et al. (2009). The electron transfer system of syntrophically grown Desulfovibrio vulgaris. J. Bacteriol. 191 5793–5801 10.1128/JB.00356-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker C. B., Redding-Johanson A. M., Baidoo E. E., Rajeev L., He Z., Hendrickson E. L., et al. (2012). Functional responses of methanogenic archaea to syntrophic growth. ISME 6 2045–2055 10.1038/ismej.2012.60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner O., Gujer W. (1986). A multispecies biofilm model. Biotechnol. Bioeng. 28 314–328 10.1002/bit.260280304 [DOI] [PubMed] [Google Scholar]
- Wei H., Yang Z., Lux R., Zhao M., Wang J., He X., et al. (2012). Direct visualization of the interaction between pilin and exopolysaccharides of Myxococcus xanthus with eGFP fused PilA protein. FEMS Microbiol. Lett. 326 23–30 10.1111/j.1574-6968.2011.02430.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilke C. R., Chang P. (1955). Correlation of diffusion coefficients in dilute solutions. AIChE J. 1 264–270 10.1002/aic.690010222 [DOI] [Google Scholar]
- Xie X., Johnson T. M., Wang Y., Lundstrom C. C., Ellis A., Wang X., et al. (2013). Mobilization of arsenic in aquifers from the Datong Basin, China: evidence from geochemical and iron isotopic data. Chemosphere 90 1878–1884 10.1016/j.chemosphere.2012.10.012 [DOI] [PubMed] [Google Scholar]
- Xu K. D., McFeters G. A., Stewart P. S. (2000). Biofilm resistance to antimicrobial agents. Microbiology 146 547–549. [DOI] [PubMed] [Google Scholar]
- Yeoh H. T., Bungay H. R., Krieg N. R. (1968). A microbial interaction involving combined mutualism and inhibition. Can. J. Microbiol. 14 491–492 10.1139/m68-083 [DOI] [PubMed] [Google Scholar]
- Zijnge V., van Leeuwen M. B. M., Degener J. E., Abbas F., Thurnheer T., Gmür R., et al. (2010). Oral biofilm architecture on natural teeth. PLoS ONE 5:e9321 10.1371/journal.pone.0009321 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.