Figure 2. Comparative analysis of the gene expression programs in human and mouse samples.
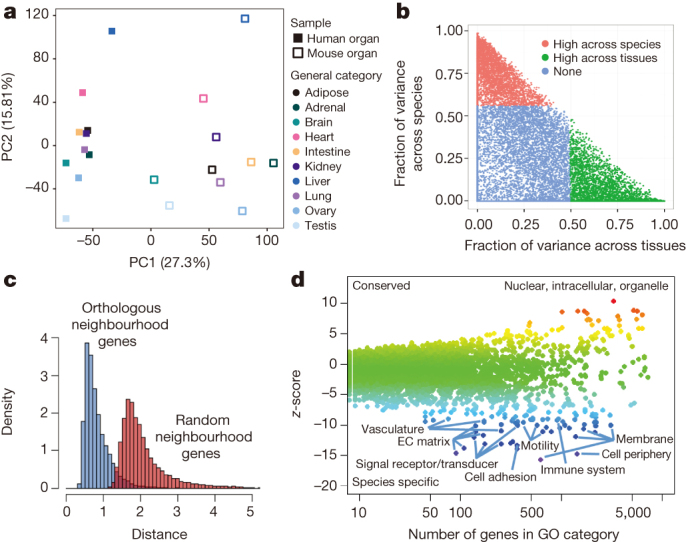
a, Principal component analysis (PCA) was performed for RNA-seq data for 10 human and mouse matching tissues. The expression values are normalized across the entire data set. Solid squares denote human tissues. Open squares denote mouse tissues. Each category of tissue is represented by a different colour. b, Gene expression variance decomposition (see Methods) estimates the relative contribution of tissue and species to the observed variance in gene expression for each orthologous human–mouse gene pair. Green dots indicate genes with higher between-tissue contribution and red dots genes with higher between-species contributions. c, Neighbourhood analysis of conserved co-expression (NACC) in human and mouse samples. The distribution of NACC scores for each gene is shown. d, A scatter plot shows the average of NACC score over the set of genes in each functional gene ontology category. Highlighted are those biological processes that tend to be more conserved between human and mouse and those processes that have been less conserved (see Supplementary Table 21 for list of genes).
