Figure 6. Human GWAS hits when mapped onto mouse genome are associated with specific chromatin states.
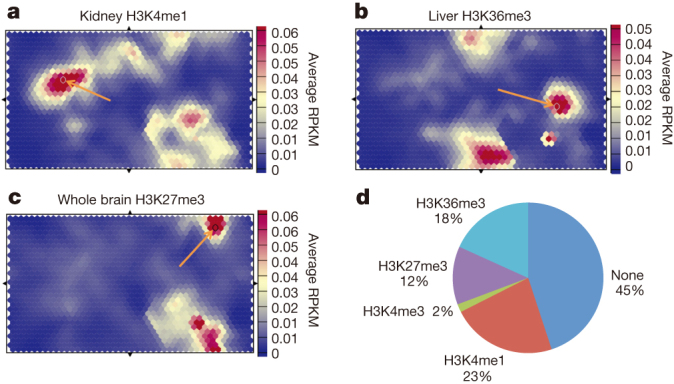
a, A self-organization map of histone modification H3K4me1 shows association between kidney H3K4me1 state and specific GWAS hits associated with urate levels (Methods). b, Liver-specific H3K36me3 unit shows enrichment in GWAS hits related to cholesterol, alcohol dependence and triglyceride levels. c, Brain-specific H3K27me3 high unit shows enrichment in GWAS SNPs associated with neurological disorders. d, Characterization of every unit with statistically significant GWAS enrichments in terms of highest histone modification signal in at least one sample. Units with no signal in top 100 map units for every histone modification are listed as none. RPKM, reads per kilobase per million reads mapped.
