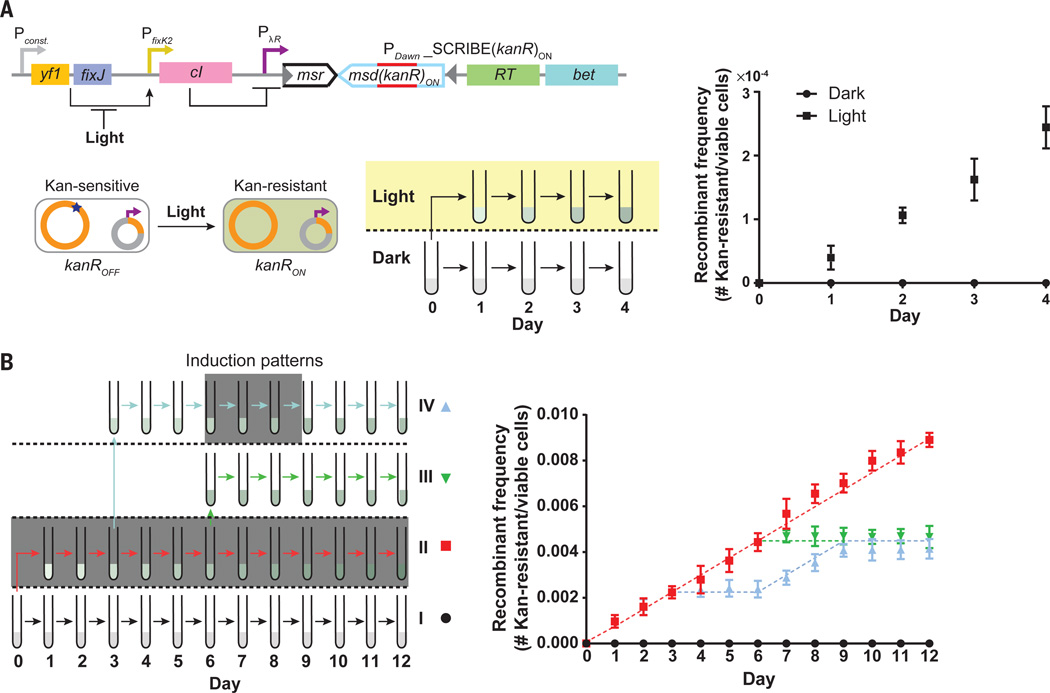
Fig. 4. Optogenetic genome editing and analog memory for long-term recording of input signal exposure times in the genomic DNA of living cell populations.
(A) We coupled expression of SCRIBE(kanR)ON to an optogenetic system (PDawn). The yf1/fixJ synthetic operon was expressed from a constitutive promoter – its products cooperatively activate the PfixK2 promoter, which drives lambda repressor (cI) expression, which subsequently represses the SCRIBE(kanR)ON cassette. Light inhibits the interaction between yf1 and fixJ, leading to the generation of ssDNA(kanR)ON and Beta expression, and thus the conversion of kanROFF to kanRON. Cells harboring this circuit were grown overnight at 37°C in the dark, diluted 1:1000, and then incubated for 24 h at 30°C in the dark (no shading) or in the presence of light (yellow shading). Subsequently, cells were diluted by 1:1000 and grown for another 24 h at 30°C in the dark or in the presence of light. The dilution/regrowth cycle was performed for four consecutive days. The kanR allele frequencies in the populations were determined by sampling the cultures after each 24-hour period. (B) SCRIBE analog memory records the total time exposure to a given input, regardless of the underlying induction pattern. Cells harboring the circuit shown in Fig. 1C were grown in four different patterns (I-IV) over a twelve-day period, where induction by IPTG (1 mM) and aTc (100 ng/mL) is represented by dark gray shading. At the end of each 24 h incubation period, cells were diluted by 1:1000 into fresh media. The number of Kan-resistant cells in the cultures was determined at the end of each day. Dashed lines represent the recombinant allele frequencies predicted by the model (see Supplementary Materials). Error bars indicate the standard error of the mean for three independent biological replicates.