Figure 1. Roles of enhancers in development, signaling events and diseases.
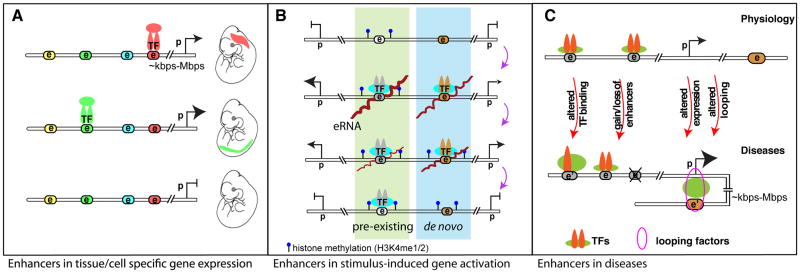
A. Differential enhancer binding patterns dictate temporal and spatial gene regulations during development. Colored nodes with “e” indicate potential enhancer elements, which can be activated during specific developmental windows upon recruitment of certain transcription factors (TF) for coordinated tissue or cell-type specific gene expression patterns. Tissue specific enhancers are color coordinated. Relative expression levels are denoted by the proportional sizes of the arrowheads. “e”, “p” stand for enhancer and promoter, respectively; double-line represents linear genomic distance between enhancer and promoter that can range from several kilo base pairs (kbps) to mega base pairs (Mbps). B. During gene transcriptional activation events in responding to differentiation or environmental cues (e.g. LPS (lipopolysaccharide) stimulation in macrophages), a group of pre-existing enhancers (highlighted green) bearing mono- or di-methylation of histone lysine 4 (H3K4me1/2) modification is readily activated. A small subset of enhancers is generated de novo in response to LPS stimulation resulting in deposition of H3K4me1/2 enhancer marks [Kaikkonen 2013]. This group of enhancers, highlighted in blue, will be readily activated upon repeated stimulation [Otsuni 2013]. The order from the top – stimulus activation – to the bottom - restoration to resting state – indicates the sequential cascade of events; the distance between two histone methylation represents nucleosome spacing and chromatin accessibility; and brown lines on “e” indicates eRNAs. C. Dysregulated enhancer functions are involved in human diseases, as exemplified in breast cancer and coronary artery disease (CAD) [5, 78, 79]. Genetic variation in enhancer (denoted as e’) could alter TF binding, gain or loss of functional enhancers, resulting in differential gene regulation. This is also often paralleled by differential chromosomal looping. Some of these scenarios can be directly linked to disease-associated genetic variants that occur at the DNA level of enhancers (e.g. single nucleotide polymorphism, insertion, deletion, and copy number variants etc.). The sizes of arrowheads of genes or symbols of TFs are proportional to their expression levels or binding intensities, respectively.
