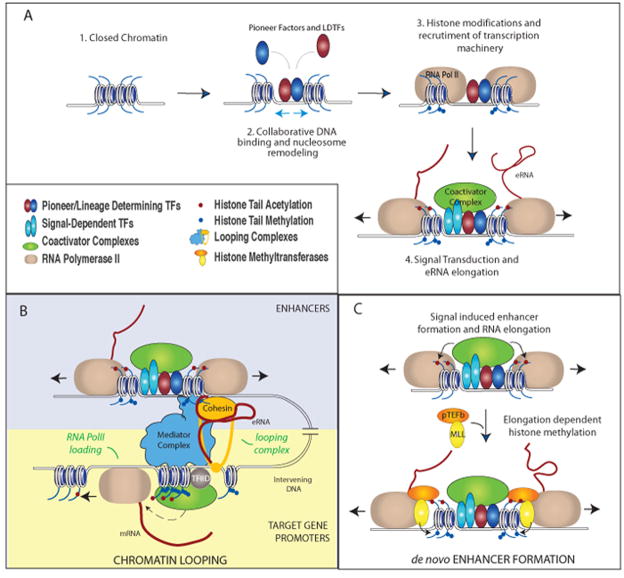
Figure 3. Proposed functional mechanisms of enhancer transcription and transcripts.
A. Collaborative binding of pioneer or lineage determining transcription factors LDTFs (red and dark blue) leads to nucleosome remodeling, increased chromatin accessibility, histone modifications (mono- and di-methylation on H3K4, blue circles on histone tails), and assembly of basal transcription machinery. Upon stimulation, signal-dependent transcription factors (SDTFs, light blue) bind to recognition sequences at enhancers and recruit coactivator complexes (green). This leads to further epigenetic modifications (e.g. histone acetylation, red circles on histone tails) and transcriptional activation of the enhancer. B–C. The enhancer transcription and transcripts, under different circumstances, may have independent functional roles. B. (A proposed model) eRNA functionally contributes to enhancer-mediated coding gene expression. Signal dependent activation of enhancers leads to increased production of eRNAs, which interacts with looping factors (e.g. cohesin complex) and facilitates/stabilizes chromosomal looping between enhancer and the promoter(s) of cognate target gene(s). eRNA mediates the loading of RNA PolII, and likely the transcription initiation complex (denoted by TFIID) at promoter of target gene [Mousavi 2013]. Whether chromosomal looping facilitates RNA PolII loading is unknown. C. Transcription elongation is required for the deposition of di-methylation of histone lysine 4, or H3K4me2, also a histone mark for enhancer, during signal-dependent activation of de novo enhancers. The coactivator complexes generate acetylation of histones, which recruits the positive transcription elongation factor (pTEFb, orange) to promote elongation of RNA Pol II. Subsequently, the elongating Pol II cargos histone methyl transferases, Mixed Lineage Leukemia complexes (MLLs), for di-methylation deposition on H3K4 at enhancers [Kaikkonen, 2013].