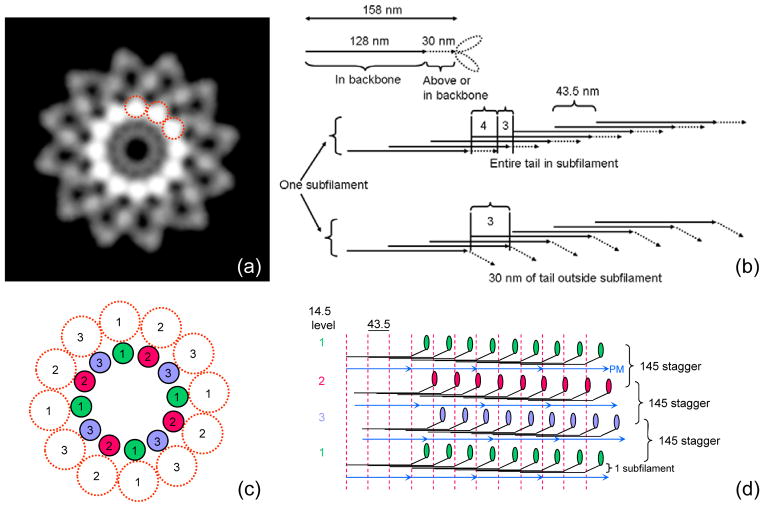
Figure 5. Backbone structure in Limulus thick filament.
(a) Density projection of one complete repeat of reconstruction (43.5 nm) along filament axis (protein white). The highest protein density is in a ring of twelve rods, about 4 nm in diameter (three have been circled), running parallel to the filament axis. Each represents one subfilament. Lower density at higher radius represents myosin heads. (b) Schematic diagram showing number of tails in cross-section of a subfilament, assuming a tail length of 158 nm (see text) and that molecules within a subfilament are staggered by 43.5 nm.24 Top: entire tail is in subfilament; subfilament thus has 4 tails in cross-section for ~ 2/3 of each repeat. Bottom: 30 nm of tail is outside subfilament; subfilament thus has 3 tails in cross-section along almost entire repeat. (c), (d) Hypothetical backbone structure showing how paramyosin molecules ~ 130 nm long might fit into the filament core, one associated with each subfilament every 3×43.5 nm. Numbers indicate relative axial levels of subfilaments and paramyosin (in increments of 14.5 nm). Molecules in each subfilament, and in subfilaments at the same level, have the same color. In this speculative model, paramyosins (assumed diameter 2 nm) are not quite close enough to interact. Such interaction, expected in any plausible model of the filament backbone, could occur, for example, if the coiled-coil diameter were slightly larger, or the filament dimensions slightly smaller than assumed. PM = paramyosin.