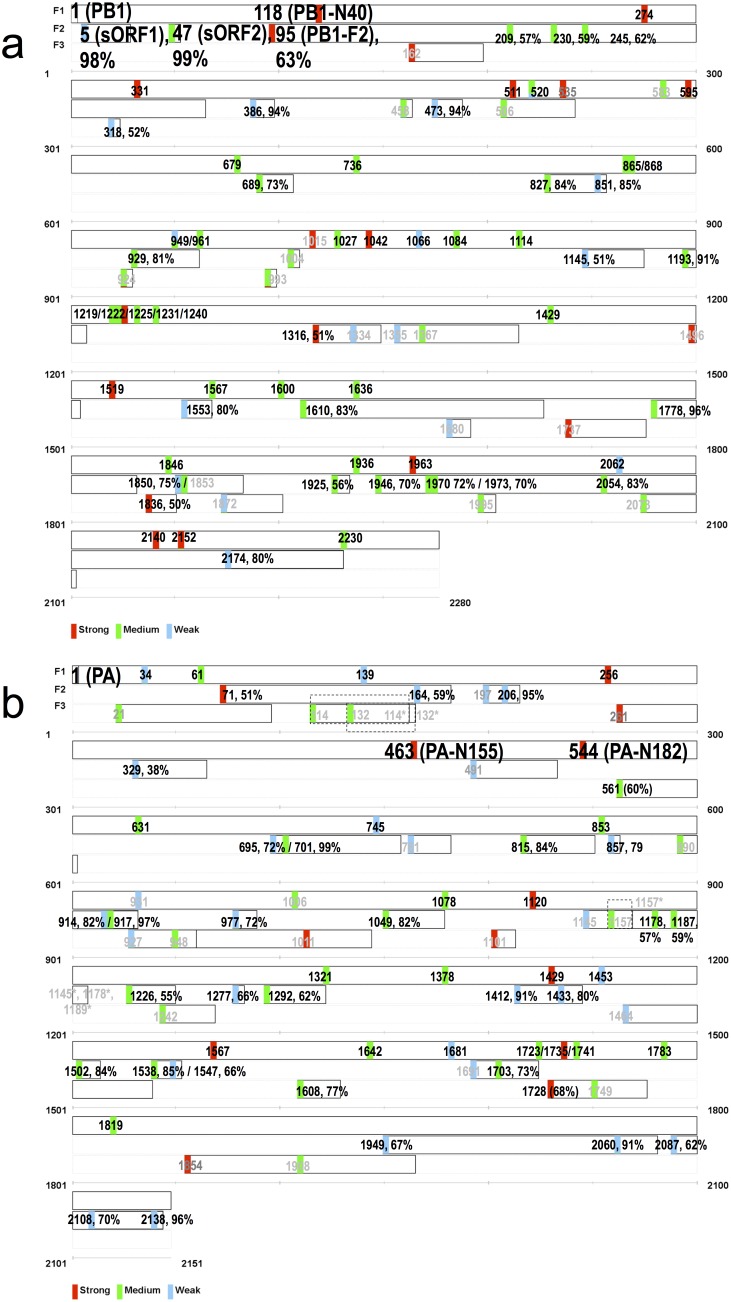
Figure 1. ORF maps of PB1 and PA genes.
a) Eighty-one putative ORFs of PB1 gene are graphed in 3 forward-reading frames: F1, F2 and F3. Each colored box represents the start codon of an ORF, with red, green and blue indicating strong, medium and weak Kozak sequences, respectively. Numbers from 1 are used by each start codon to indicate positions. For ORFs with mixed populations of different Kozak strengths, multiple colors are used to represent the population proportions. For example, the ORF at position 520 is 74% green and 26% blue. F2 and F3 ORFs of major prevalence (>50%) are additionally labeled by their frequencies. ORFs of lower prevalence (<95% for F1, <50% for F2/F3) are grey-labelled. Each ORF ends with a solid line. For F2 or F3 ORFs of various ORF sizes, the corresponding dominant length is used to close the ORF. Four previously reported PB1 ORFs, PB1-F2, PB1-N40, sORF1 and sORF2, are bold-highlighted. b) Seventy-seven putative ORFs of the PA gene. The previously reported PA-N155 and PA-N182 are bold-highlighted at positions 463 and 544, respectively. Note that some ORFs overlap, such as those at positions 114 and 132. In such cases, 114* and 132*, in grey, are used to mark the end of the individual ORF, starting from position 114 and 132, respectively.