Abstract
Papillary thyroid cancer (PTC) is the most common epithelial thyroid tumor, accounting for more than 80% of all thyroid cancers. Although PTC shows an indolent character and excellent prognosis, patients with aggressive characteristics are more likely to have a disease recurrence and die in the end. The aim of this study was to analyze BRAFV600E mutation and methylation levels of CpG sites in the promoters of CDH1, DAPK, RARβ and RUNX3 genes in a cohort of PTCs, and investigate their association with tumor recurrence. In this study, we used pyrosequencing method to individually quantified methylation levels at multiple CpG sites within each gene promoter, and detect BRAFV600E mutation in 120 PTCs and 23 goiter tissues as normal control. Moreover, appropriate cut-off values for each CpG site were set up to predict disease recurrence. Our data showed that overall average methylation levels of CDH1 and RUNX3 genes were significantly higher in PTCs than that in control subjects. Conversely, overall average methylation levels of DAPK promoter were significantly lower in PTCs than that in control subjects. Moreover, BRAFV600E mutation and overall average methylation levels of all these genes were not significant difference between recurrent and non-recurrent cases. However, we found that hypermethylation of RUNX3 at CpG sites -1397, -1406, -1415 and -1417 significantly increased the risk of of disease recurrence by using appropriate site-specific cut-off values. Collectively, our findings suggest RUNX3 site-specific hypermethylation may offer value in predicting or monitoring postoperative recurrence of PTC patients.
Keywords: Papillary thyroid cancer (PTC), tumor recurrence, RUNX3, promoter hypermethylation, pyrosequencing
Introduction
Thyroid cancer is the most common endocrine malignancy, the incidence of which has increased continuously in recent years. The most common histological type is particularly papillary thyroid cancer (PTC), which accounts for more than 80% of all thyroid cancers [1,2]. Although the majority of PTCs can be efficiently cured by surgical and radioiodinated therapy and have an excellent prognosis, recurrent disease develops in about 20% to 30% of patients with lymph node metastasis, and ~7% of patients die from progressive disease within 10 years of diagnosis [3]. Therefore, there is pressing need to develop an effective strategy for predicting or monitoring tumor recurrence before or after initial surgery. It would be clinically important for improving survival and quality of life in patients with PTC.
It is well known that BRAFV600E mutation is the most prominent oncogenic event in PTC, which results in constitutively activated BRAF kinase [4]. It plays a critical role in PTC tumorigenesis through driving Ras/Raf/MEK/ERK (MAPK) signaling [4,5]. However, the association of BRAFV600E mutation with PTC recurrence remains controversial [6,7]. In addition, DNA methylation, one of epigenetic changes, is also the most extensively studied in diverse human cancers and can be used as potential biomarkers for cancer diagnosis, including PTC [8]. Generally, DNA methylation as an important hallmark of cancer cells is one of the major mechanisms to inactivate tumor suppressor genes and seriously affect their function, contributing to carcinogenesis [9,10]. It has been demonstrated that promoter hypermethylation of certain genes is associated with disease recurrence in diverse cancers, including bladder cancer, non-small cell lung cancer, prostate cancer and hepatocellular carcinoma [11-14].
In this study, we investigated methylation levels of CDH1, DAPK, RARβ and RUNX3 genes and BRAFV600E mutation in a cohort of PTC samples by using pyrosequencing method, and attempted to discover CpG site-specific hypermethylation for accurately predicting disease recurrence and determine the association of BRAFV600E mutation with PTC recurrence.
Material and methods
Patients and tissue samples
With the approval of our institutional review board and human ethics committee, a total of 120 paraffin-embedded PTCs and 23 goiter tissues used as control subjects were randomly obtained at the First Affiliated Hospital of Xi’an Jiaotong University School of Medicine. None of these patients received chemotherapy and radiotherapy before the surgery. Informed consent was obtained from each patient before the surgery. All paraffin-embedded tissues were examined and agreed upon by a senior pathologist at Department of Pathology of the Hospital. Clinicopathologic data, including gender, age, lymph node metastasis, tumor stage and recurrence, was collected retrospectively, and was summarized in Supplementary Table 1.
DNA isolation
Genomic DNA was extracted from isolated tissues as previously described [19]. Briefly, after a treatment for 12 h at room temperature with xylene to remove paraffin, the tissues were digested with 1% sodium dodecyl sulfate (SDS) and 0.5 mg/ml proteinase K at 48°C for 48 h, with addition of several spiking aliquots of concentrated proteinase K to facilitate digestion. Genomic DNA was then isolated from the digested tissues using standard phenol/chloroform protocol, and was dissolved in distilled water and stored at -80°C until use.
Sodium bisulfite treatment
DNA from thyroid tissues and cell lines was subjected to bisulfite treatment as described previously [15]. Briefly, a final volume of 20 μl of H2O containing ~2 μg genomic DNA, 10 μg salmon sperm DNA, and 0.3 M NaOH was incubated at 50°C for 20 min to denature the DNA, followed by incubation in a 500 μl reaction mixture freshly prepared containing 3 M sodium bisulfite (Sigma, Saint Louis, MO) and 10 mM hydroquinone (Sigma, Saint Louis, MO) at 70°C for 3 h. The DNA was then purified by a Wizard DNA Clean-Up System (Promega Corp., Madison, WI) according to the instructions of the manufacturer, followed by ethanol precipitation, and resuspension in 50 μl of deionized H2O after vacuum drying for DNA methylation analysis. Bisulfite-treated DNA samples were stored at -80°C until use.
DNA methylation analysis using pyrosequencing
We used bisulfite pyrosequencing assay to quantitatively analyze methylation levels of 4 genes, including CDH1, DAPK, RARβ and RUNX3. The amplifying and sequencing primers were designed using PyroMark Assay design software ver. 2.0 (Qiagen, Valencia, CA), and the sequences were presented in Supplementary Table 2. The PCR was conducted in a volume of 30 μl with ~30 ng bisulfite-converted DNA, 1×PCR buffer, 3 mM MgCl2, 300 μM each of deoxynucleotide triphosphate mixture (dATP, dCTP, dGTP and dTTP), 300 nM forward and reverse primers, and 1.0 U of platinum Taq DNA polymerase (Invitrogen Technologies, Inc., CA). The amplification was performed in a Thermal cycler (Bio-Rad Laboratory, Inc., CA) as follows: after a 5 min denaturation at 95°C, the reaction was run for 40 cycles, each comprising 45 s of denaturing at 95°C, 45 s of annealing at variable temperatures according to the primers, and 45 s of extension at 72°C, with an extension at 72°C for 5 min as the last step. The PCR products were electrophoresed on a 1.2% agarose gel and visualized under UV illumination using an ethidium bromide stain.
A biotin-labeled primer was used in the PCR in order to purify the final product from the mix by using streptavidin-coated Sepharose beads (10058558; GE Healthcare, Milwaukee, WI). Sequencing primer was then annealed to the purified single-stranded PCR product, and pyrosequencing was performed on a PyroMarkQ24 System (Qiagen, Germany) according to manufacturer’s instructions. To provide an internal control for total bisulfite conversion, a non-CG cytosine was included in the region targeted for pyrosequencing where possible. Target CpG sites were evaluated by using the instrument software (PSQ24MA 2.0.6, Qiagen), which converts pyrograms to numerical values for peak heights and calculates the proportion of methylation at each base as a C/T ratio.
Detection of BRAFV600E mutation by pyrosequencing
A 228-bp fragment from exon 15 of the BRAF gene spanning the hotspot mutation site at codon 600 was analyzed by pyrosequencing method as described previously [16].
Statistical analysis
The output data include site-specific percent methylation for each CpG in the investigated regions as well as overall average percent methylation, which is the average of the CpG methylation levels within the promoter. Overall or site-specific methylation levels of each gene between recurrent and non-recurrent groups were compared using the independent sample t-test or non-parametric Wilcoxon’s Rank Sum test according to the distribution type of the data. Similar statistical strategy was used to evaluate the association of BRAF mutation with tumor recurrence. To predict disease recurrence, we set up site-specific cut-off values by constructing receiver operator characteristic (ROC) curves using MedCalc Software (MedCalc Software bvba, Belgium). Site-specific sensitivity and specificity, positive and negative predictive values were calculated using standard formulas. Univariate and multivariable logistic regression analyses were performed to estimate the odds ratios (ORs) of clinicopathologic characteristics and disease recurrence according to site-specific methylation levels, independent of gender, age, lymph node metastasis and TNM stage. Kaplan-Meier survival analysis and standard log-rank test were used to evaluate the effect of site-specific methylation levels on tumor recurrence. All statistical analyses were performed using the SPSS statistical package (11.5, Chicago, IL, USA). P values < 0.05 were considered significant.
Results
Overall average methylation levels of CDH1, DAPK, RUNX3 and RARβ genes in PTCs
We evaluated promoter methylation of CDH1, DAPK, RUNX3 and RARβ genes in 120 PTCs and 23 goiter tissues by using pyrosequencing method. As shown in Figure 1, overall average methylation levels of CDH1, RUNX3 and RARβ were higher in PTCs than in control subjects. In contrast, overall average methylation levels of DAPK were lower in PTCs than in control subjects. Among these genes examined, statistical significances were found in CDH1 (P = 0.03), DAPK (P = 0.03), and RUNX3 (P < 0.001). Given the importance of promoter methylation in thyroid tumorigenesis, the association of promoter methylation of these genes with clinicopathological features, including gender, age, lymph node metastasis, TNM stage and tumor recurrence, was explored in a cohort of PTCs. However, our data showed that overall average methylation levels of these four genes did not significantly correlate with any clinicopathological variables in PTCs (data not shown). Although overall average methylation levels of these four genes were a little bit higher in recurrent cases than in non-recurrent cases, the differences did not reach statistical significance (Figure 2).
Figure 1.
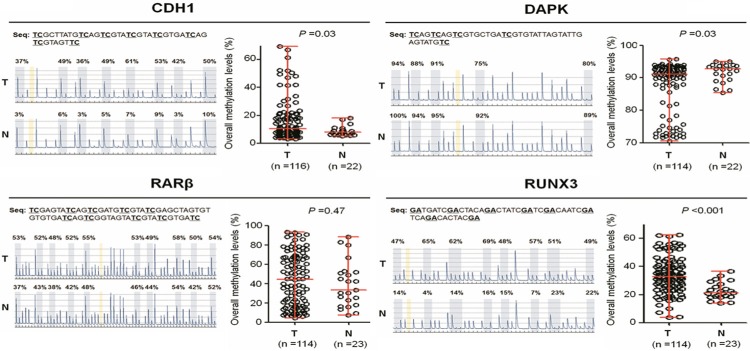
Analysis of overall methylation levels of four tumor suppressor genes in PTC samples using pyrosequencing assay. Pyrosequencing method was performed as described in Methods. Representative pyrograms show methylation levels of the indicated CpG sites in gene promoter (left panel). The average methylation level of each gene is represented in the right panel. Horizontal lines represent the median and the 25th to 75th percentile. T, PTC tissues; N, goiter tissues. P values < 0.05 were considered significant.
Figure 2.
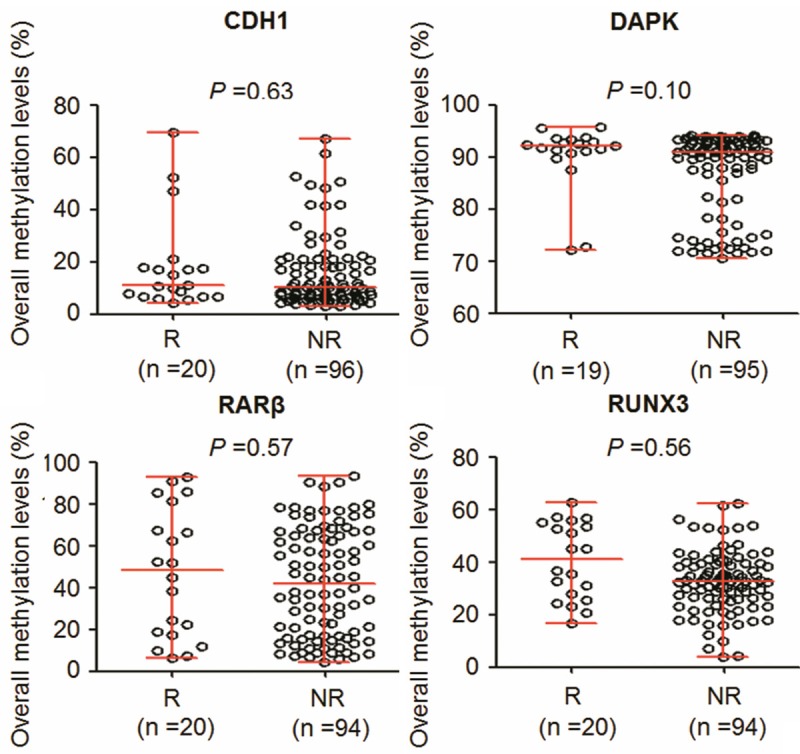
Association of overall methylation levels of these four genes with tumor recurrence. Shown were scatterplots of average methylation levels of each gene in recurrent (R) and non-recurrent (NR) cases. Horizontal lines represent the median and the 25th to 75th percentile.
Detection of BRAFV600E mutation in PTCs using pyrosequencing
We used pyrosequencing assay to test BRAFV600E mutation in a total of 120 PTCs. In these PTCs, 92 of 120 (76.7%) cases were positive for BRAF mutation. As expected, all control subjects were negative for BRAF mutation. Given that previous studies have revealed an association of BRAFV600E mutation with promoter methylation of tumor suppressor genes in thyroid cancer [16-20], we thus tested the association of methylation levels of these genes with this mutation found in these PTCs. Of these four methylation markers investigated, BRAF mutation was only positively associated with overall average methylation levels of RARβ (P = 0.01) by Wilcoxon test analysis (Figure 3). In addition, we investigated the relationship between BRAF mutation and disease recurrence. The results showed that this mutation was not significantly associated with tumor recurrence (P = 0.629).
Figure 3.
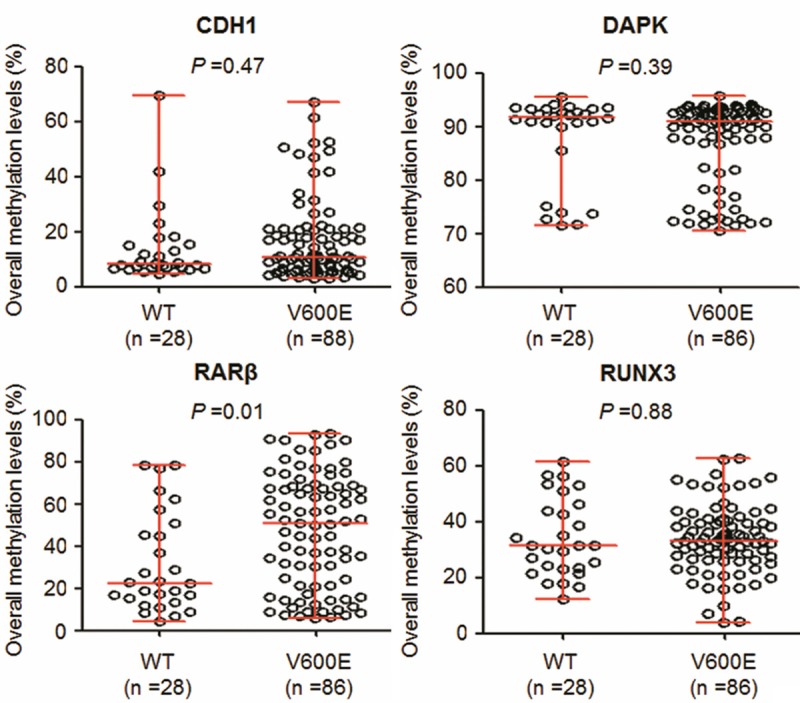
Association of BRAF mutation with overall methylation levels of these four genes in PTCs. Shown were scatterplots of overall methylation levels of each gene in BRAF mutation-negative (WT) and -positive (V600E) cases. Horizontal lines represent the median and the 25th to 75th percentile. P values < 0.05 were considered significant.
Association of RUNX3 site-specific methylation with tumor recurrence
The site-specific percent methylation of each of multiple CpG sites within CDH1, DAPK, RUNX3 and RARβ promoters was measured and investigated their association with clinicopathologic characteristics in PTCs. The results showed that site-specific methylation levels in these gene promoters did not significantly correlate with most of clinicopathologic variables (data not shown). However, we found that methylation levels of RUNX3 at CpG sites -1397, -1406, -1415 and -1417 were associated with tumor recurrence. Methylation levels at these CpG sites were much higher in recurrent cases than in non-recurrent cases, particularly at the latter three sites (P = 0.04 at CpG site -1406; P = 0.01 at CpG site -1415; P < 0.001 at CpG site -1417) (Figure 4).
Figure 4.
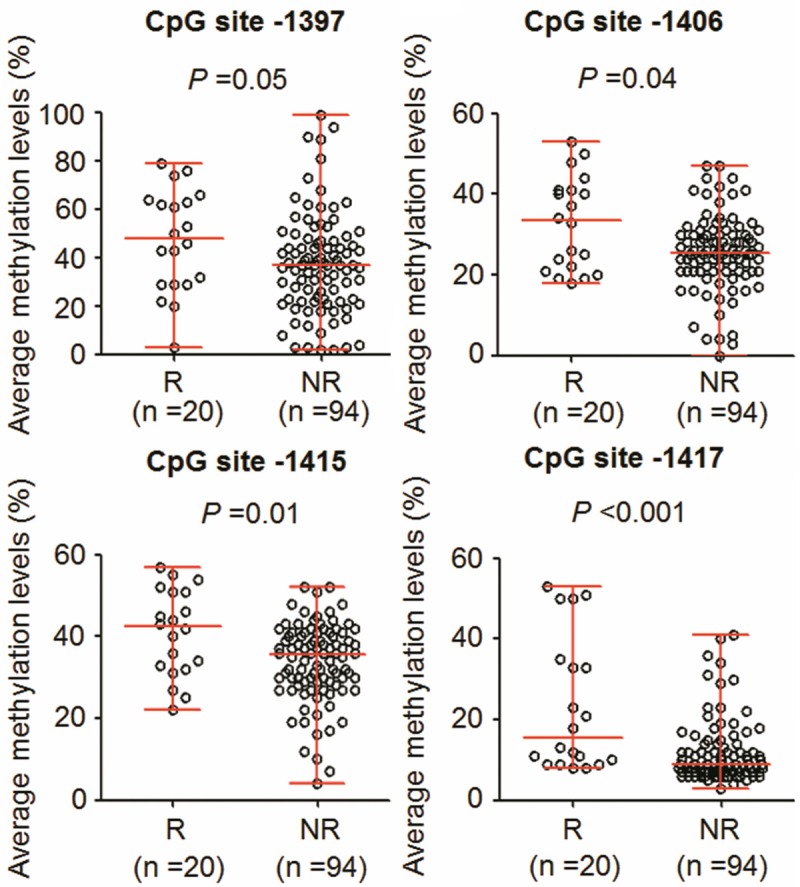
Association of RUNX3 site-specific methylation levels with tumor recurrence. Shown were scatterplots of RUNX3 site-specific methylation levels in recurrent (R) and non-recurrent (NR) cases. Horizontal lines represent the median and the 25th to 75th percentile. P values < 0.05 were considered significant.
Next, we set up appropriate cut-off values for these four CpG sites to accurately predict tumor recurrence by employing receiver operator characteristic (ROC) curves. ROC curve is graphic presentation of the relationship between both sensitivity and specificity and it helps to decide the optimal model through determining the best threshold for a diagnostic/predictive test [21]. The area under the curve (AUC) of the ROC curves provides a way to measure the accuracy of a diagnostic/predictive test. The large area, the more accurate the test is [21]. As shown in Table 1 and Figure 5, all these four CpG sites showed high AUC of the ROC curves, ranging from 0.64 to 0.75. Importantly, RUNX3 site-specific hypermethylation was associated with a significantly increased risk of tumor recurrence (CpG site -1397: OR = 4.50, 95% CI = 1.53-13.26, P = 0.011; CpG site -1406: OR = 5.33, 95% CI = 1.56-18.18, P = 0.013; CpG site -1415: OR = 5.71, 95% CI = 2.01-16.24, P = 0.001; CpG site -1417: OR = 12.12, 95% CI = 3.11-47.16, P = 0.000). These findings suggest that RUNX3 site-specific hypermethylation may be a dangerous factor for PTC recurrence. To evaluate the value of RUNX3 site-specific methylation in predicting PTC recurrence, we calculated their sensitivity and specificity by the use of the same cut-off values. Also shown in Table 1, although the sensitivities of these four CpG sites were not too high, ranging 30.0 to 50.0%, they showed the excellent specificities, ranging from 85.1 to 95.7%, and the negative predictive values of 86.1 to 88.9%. Taken together, RUNX3 site-specific hypermethylation has predictive value for postoperative recurrence of PTC patients.
Table 1.
The values of RUNX3 site-specific hypermethylation in predicting PTC recurrence
| CpG sites | AUC1 | Cut-off values | OR (95% CI)2 | P values | Sensitivity (%) | Specificity (%) | PPV (%)3 | NPV (%)4 |
|---|---|---|---|---|---|---|---|---|
| -1397 | 0.64 | 59.0% | 4.50 (1.53-13.26) | 0.011 | 40.0 (8/20) | 87.1 (81/93) | 40.0 (8/20) | 87.1 (81/93) |
| -1406 | 0.65 | 40.5% | 5.33 (1.56-18.18) | 0.013 | 30.0 (6/20) | 92.6 (87/94) | 46.2 (6/13) | 86.1 (87/101) |
| -1415 | 0.69 | 42.5% | 5.71 (2.01-16.24) | 0.001 | 50.0 (10/20) | 85.1 (80/94) | 41.7 (10/24) | 88.9 (80/90) |
| -1417 | 0.75 | 32.0% | 12.12 (3.11-47.16) | 0.000 | 35.0 (7/20) | 95.7 (90/94) | 63.6 (7/11) | 87.4 (90/103) |
AUC (area under the curve).
OR: odds ratio with 95% confidence interval (CI).
PPV, positive predictive value.
NPV, negative predictive value.
Figure 5.
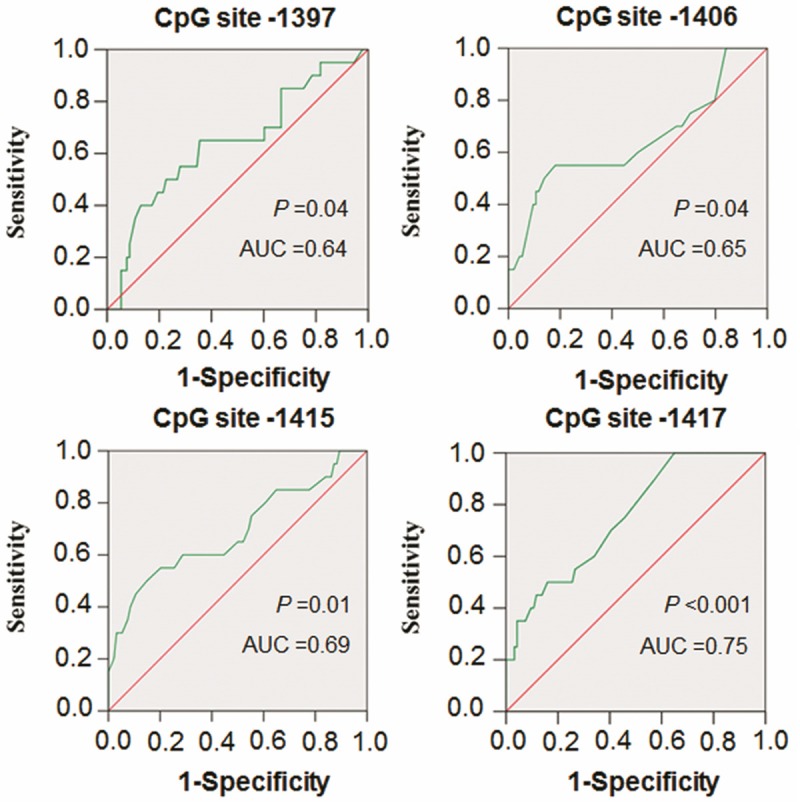
Receiver operating characteristic (ROC) curves for four CpG sites in the RUNX3 promoter. All recurrent and non-recurrent PTCs for which there was complete DNA methylation data were used for the analysis. The ROC curves plot sensitivity and 1-specificity. Areas under the curve (AUC) and P values were shown in the graph.
The prognostic value of RUNX3 site-specific hypermethylation for PTC recurrence was further demonstrated as dramatically reduced disease-free probability by Kaplan-Meier analysis using the same cut-off values (CpG -1397: 95.1 months vs. 125.2 months on average, P = 0.20; CpG -1406: 65.7 months vs. 134.0 months on average, P < 0.001; CpG -1415: 80.3 months vs. 138.1 months on average, P = 0.007; CpG -1417: 59.3 months vs. 135.6 months on average, P = 0.006) (Figure 6). We next want to explore whether the prognostic value of these site-specific hypermethylation is attributable to its relationship with other potential clinicopathologic confounding factors, including gender, age, lymph node metastasis and TNM stage, or whether they contribute independently to prognosis. Thus, the independent relationship of these site-specific hypermethylation with disease-free survival was studied using Cox multivariate repression analysis. The results indicated that RUNX3 site-specific hypermethylation is a predictor of poor prognosis for PTC recurrence as an independently variable with respect to gender, age, lymph node metastasis and TNM stage (Table 2).
Figure 6.
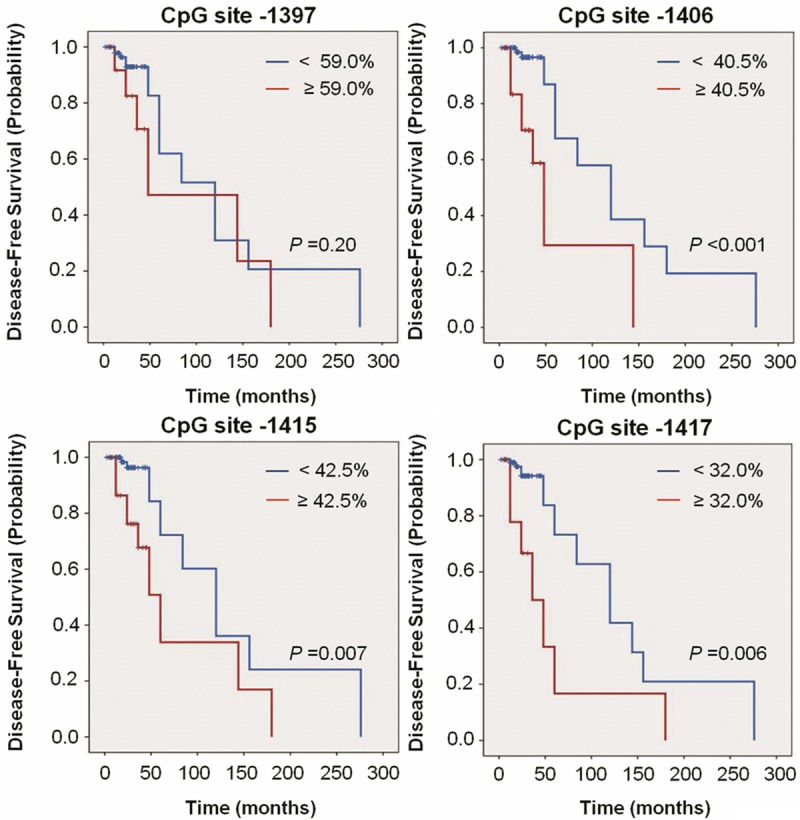
Kaplan-Meier estimate of PTC disease-free probability in patients with different RUNX3 site-specific methylation levels. Short vertical lines indicate censored observations (months of follow-up for those who have no had recurrence/persistence).
Table 2.
The prognostic value of clinicopathological factors and RUNX3 site-specific hypermethylation in multivariate Cox regression analysis
| Variables | RUNX3 gene | |||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| CpG site -1397 | CpG site -1406 | CpG site -1415 | CpG site -1417 | |||||
|
|
|
|
|
|||||
| Hazard Ratio (HR) (95% CI) | P | Hazard Ratio (HR) (95% CI) | P | Hazard Ratio (HR) (95% CI) | P | Hazard Ratio (HR) (95% CI) | P | |
| Methylation levels | ||||||||
| < cut-off values | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | ||||
| ≥ cut-off values | 5.83 (1.66-20.44) | 0.006 | 16.92 (4.42-64.76) | 0.000 | 8.06 (2.47-26.27) | 0.001 | 12.69 (2.94-54.69) | 0.001 |
| Gender | ||||||||
| Female | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | ||||
| Male | 0.53 (0.13-2.26) | 0.39 | 0.94 (0.24-3.72) | 0.93 | 0.75 (0.20-2.85) | 0.67 | 1.15 (0.29-4.57) | 0.85 |
| Age | ||||||||
| ≤ 40 | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | ||||
| > 40 | 0.11 (0.01-0.93) | 0.04 | 0.26 (0.04-1.84) | 0.18 | 0.21 (0.03-1.56) | 0.13 | 0.27 (0.03-2.23) | 0.23 |
| Lymph node metastasis | ||||||||
| No | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | ||||
| Yes | 2.72 (0.66-11.24) | 0.17 | 4.60 (1.08-19.72) | 0.04 | 3.81 (0.91-16.00) | 0.07 | 3.61 (0.96-13.68) | 0.06 |
| TNM Stage | ||||||||
| < III | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | ||||
| ≥ III | 5.11 (0.75-34.93) | 0.09 | 1.82 (0.34-9.61) | 0.48 | 1.67 (0.31-9.07) | 0.55 | 0.71 (0.10-4.95) | 0.73 |
Discussion
It is well known that patients with PTC have excellent prognosis, but 5%-40% of cases have persistent or recurrent disease even after curative surgery [22,23]. Although radioiodine treatment may reduce recurrence, 131I ablation was effective in only 10% of high-risk PTC patients according to the AMES (age, metastasis to distant sites, extrathyroidal invasion, and size) classification, and was not effective in 30% of low-risk PTC patients [24-26]. Thus, early predicting the risk of cancer recurrence, such as aggressive PTCs, is very important to improve clinical outcomes of patients.
Assays of serum thyroglobulin (Tg) concentrations combined with high-resolution ultrasound have been widely used for postoperative follow-up, enabling early detection of recurrence and more prompt management [27,28]. However, the utility of Tg testing is limited by autoantibody interference in 25% of patients [29,30], and the accuracy of ultrasonography has been found to vary widely, from 40% to 90%, and to be operator-dependent [31]. In recent years, a number of evidences have demonstrated the association of gene methylation with disease recurrence in diverse cancers [14,32-34], including thyroid cancer [35]. However, these studies make use of methods that qualitatively analyze particular CpG sites within gene promoter or quantitatively analyze overall methylation levels of candidate gene as well as high-cost and global screening of aberrant DNA methylation. Therefore, in this study, we attempted to use pyrosequencing method to afford a more detailed survey of methylation state and better assess the recurrence risk of postoperative PTC patients. There are several reasons to support the point that the use of pyrosequencing assay to quantitatively analyze multiple CpG sites with gene promoter may be a useful and cost-effective strategy for predicting PTC recurrence. First, promoter methylation by gene silencing of tumor-related genes, particularly tumor suppressor genes, is one of the commonest molecular events in human tumorigenesis and progression, including PTC [5,8,36]. Second, pyrosequencing is one of the most accurate methods available for quantifying DNA methylation at specific CpG sites within the target region of interest. It is a sensitive and highly reproducible method that is uniquely suited to the analysis of a small fraction of the total DNA in the clinical samples [37,38].
In this study, we used pyrosequencing method to analyze BRAFV600E mutation and promoter methylation of four tumor suppressor genes in a cohort of PTCs and explore their association with PTC recurrence. Our data showed that the prevalence of BRAF mutation was 76.7%, and was highly specific for PTCs. However, BRAF mutation was not associated with PTC recurrence, which was consistent with a previous study [7]. Methylation analysis showed that two of these four genes tested had significantly higher overall methylation levels in PTCs than in control subjects, including CDH1 and RUNX3 genes. CDH1 (also known as E-cadherin) is a transmembrane glycoprotein that is localized in the adherens junctions of epithelial cells [39]. CDH1 inactivation by promoter methylation is thought to contribute to tumor progression through increased proliferation, invasion and metastasis [39,40]. RUNX3 belongs to the RUNX family of transcription factor and acts as strong tumor suppressor activity through modulating a series of cancer-associated genes, such as p53, p21, Notch1 and p27, which is involved in the regulation of epithelial proliferation and apoptosis [41]. DNA methylation-mediated RUNX3 inactivation has also been reported in various solid tumors, including PTC and plays a critical role in PTC tumorigenesis [42].
Next, we sought to explore the association of methylation levels of these genes with PTC recurrence. Unfortunately, our data suggested that overall methylation levels of these genes did not significantly correlate with disease recurrence. However, when we further analyzed the association of CpG site-specific methylation levels in each promoter with PTC recurrence, we found that high methylation levels of RUNX3 at CpG sites -1397, -1406, -1415 and -1417 were associated with a significantly increased risk of disease recurrence. Moreover, multivariable logistic regression showed that RUNX3 site-specific hypermethylation was a significant independent predictor for PTC recurrence. Importantly, using these CpG sites with appropriate cut-off values, we were able to obtain the excellent specificities and the negative predictive values.
In summary, in this study, we investigated methylation levels of CDH1, DAPK, RARβ and RUNX3 genes and BRAFV600E mutation in a cohort of PTCs using pyrosequencing method and demonstrated the association of RUNX3 site-specific methylation levels with PTC recurrence. These observations provide strong evidences suggesting that RUNX3 site-specific hypermethylation may be potential and reliable biomarkers to predict or monitor postoperative recurrence of PTC patients.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 81171969, 81272933 and 81372217), the Fundamental Research Funds for the Central Universities, and the Ph. D. Programs Foundation of Ministry of Education of China (No. 2012021110058).
Disclosure of conflict of interest
The authors declare that they have no competing interests.
Supporting Information
References
- 1.Xing M. Molecular pathogenesis and mechanisms of thyroid cancer. Nat Rev Cancer. 2013;13:184–99. doi: 10.1038/nrc3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 3.Hundahl SA, Fleming ID, Fremgen AM, Menck HR. A National Cancer Data Base report on 53,856 cases of thyroid carcinoma treated in the U. S., 1985-1995 [see commetns] . Cancer. 1998;83:2638–48. doi: 10.1002/(sici)1097-0142(19981215)83:12<2638::aid-cncr31>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 4.Xing M. BRAF mutation in papillary thyroid cancer: pathogenic role, molecular bases, and clinical implications. Endocr Rev. 2007;28:742–62. doi: 10.1210/er.2007-0007. [DOI] [PubMed] [Google Scholar]
- 5.Xing M. Molecular pathogenesis and mechanisms of thyroid cancer. Nat Rev Cancer. 2013;13:184–99. doi: 10.1038/nrc3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim TY, Kim WB, Rhee YS, Song JY, Kim JM, Gong G, Lee S, Kim SY, Kim SC, Hong SJ, Shong YK. The BRAF mutation is useful for prediction of clinical recurrence in low-risk patients with conventional papillary thyroid carcinoma. Clin Endocrinol (Oxf) 2006;65:364–8. doi: 10.1111/j.1365-2265.2006.02605.x. [DOI] [PubMed] [Google Scholar]
- 7.Pelttari H, Schalin-Jäntti C, Arola J, Löyttyniemi E, Knuutila S, Välimäki MJ. BRAFV600E mutation does not predict recurrence after long-term follow-up in TNM stage I or II papillary thyroid carcinoma patients. APMIS. 2012;120:380–6. doi: 10.1111/j.1600-0463.2011.02844.x. [DOI] [PubMed] [Google Scholar]
- 8.Xing M. Gene methylation in thyroid tumorigenesis. Endocrinology. 2007;148:948–53. doi: 10.1210/en.2006-0927. [DOI] [PubMed] [Google Scholar]
- 9.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–92. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCabe MT, Brandes JC, Vertino PM. Cancer DNA methylation: molecular mechanisms and clinical implications. Clin Cancer Res. 2009;15:3927–37. doi: 10.1158/1078-0432.CCR-08-2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao Y, Guo S, Sun J, Huang Z, Zhu T, Zhang H, Gu J, He Y, Wang W, Ma K, Wang J, Yu J. Methylcap-seq reveals novel DNA methylation markers for the diagnosis and recurrence prediction of bladder cancer in a Chinese population. PLoS One. 2012;7:e35175. doi: 10.1371/journal.pone.0035175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harada H, Miyamoto K, Yamashita Y, Nakano K, Taniyama K, Miyata Y, Ohdan H, Okada M. Methylation of breast cancer susceptibility gene 1 (BRCA1) predicts recurrence in patients with curatively resected stage I non-small cell lung cancer. Cancer. 2013;119:792–8. doi: 10.1002/cncr.27754. [DOI] [PubMed] [Google Scholar]
- 13.Haldrup C, Mundbjerg K, Vestergaard EM, Lamy P, Wild P, Schulz WA, Arsov C, Visakorpi T, Borre M, Høyer S, Orntoft TF, Sørensen KD. DNA methylation signatures for prediction of biochemical recurrence after radical prostatectomy of clinically localized prostate cancer. J. Clin. Oncol. 2013;31:3250–8. doi: 10.1200/JCO.2012.47.1847. [DOI] [PubMed] [Google Scholar]
- 14.Kanda M, Nomoto S, Oya H, Takami H, Hibino S, Hishida M, Suenaga M, Yamada S, Inokawa Y, Nishikawa Y, Asai M, Fujii T, Sugimoto H, Kodera Y. Downregulation of DENND2D by promoter hypermethylation is associated with early recurrence of hepatocellular carcinoma. Int J Oncol. 2014;44:44–52. doi: 10.3892/ijo.2013.2165. [DOI] [PubMed] [Google Scholar]
- 15.Shi J, Zhang G, Yao D, Liu W, Wang N, Ji M, He N, Shi B, Hou P. Prognostic significance of aberrant gene methylation in gastric cancer. Am J Cancer Res. 2012;2:116–29. [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang B, Liu S, Zhang Z, Wei J, Qu Y, Wu K, Yang Q, Hou P, Shi B. Analysis of BRAF(V600E) mutation and DNA methylation improves the diagnostics of thyroid fine needle aspiration biopsies. Diagn Pathol. 2014;9:45. doi: 10.1186/1746-1596-9-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu S, Liu D, Tufano RP, Carson KA, Rosenbaum E, Cohen Y, Holt EH, Kiseljak-Vassiliades K, Rhoden KJ, Tolaney S, Condouris S, Tallini G, Westra WH, Umbricht CB, Zeiger MA, Califano JA, Vasko V, Xing M. Association of aberrant methylation of tumor suppressor genes with tumor aggressiveness and BRAF mutation in papillary thyroid cancer. Int J Cancer. 2006;119:2322–9. doi: 10.1002/ijc.22110. [DOI] [PubMed] [Google Scholar]
- 18.Hoque MO, Rosenbaum E, Westra WH, Xing M, Ladenson P, Zeiger MA, Sidransky D, Umbricht CB. Quantitative assessment of promoter methylation profiles in thyroid neoplasms. J Clin Endocrinol Metab. 2005;90:4011–8. doi: 10.1210/jc.2005-0313. [DOI] [PubMed] [Google Scholar]
- 19.Hou P, Liu D, Xing M. Genome-wide alterations in gene methylation by the BRAF V600E mutation in papillary thyroid cancer cells. Endocr Relat Cancer. 2011;18:687–97. doi: 10.1530/ERC-11-0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brait M, Loyo M, Rosenbaum E, Ostrow KL, Markova A, Papagerakis S, Zahurak M, Goodman SM, Zeiger M, Sidransky D, Umbricht CB, Hoque MO. Correlation between BRAF mutation and promoter methylation of TIMP3, RARβ2 and RASSF1A in thyroid cancer. Epigenetics. 2012;7:710–9. doi: 10.4161/epi.20524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kampfrath T, Levinson SS. Brief critical review: Statistical assessment of biomarker performance. Clin Chim Acta. 2013;419:102–7. doi: 10.1016/j.cca.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 22.Caron NR, Clark OH. Well differentiated thyroid cancer. Scand J Surg. 2004;93:261–71. doi: 10.1177/145749690409300403. [DOI] [PubMed] [Google Scholar]
- 23.Xing M, Haugen BR, Schlumberger M. Progress in molecular-based management of differentiated thyroid cancer. Lancet. 2013;381:1058–69. doi: 10.1016/S0140-6736(13)60109-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Woodrum DT, Gauger PG. Role of 131I in the treatment of well differentiated thyroid cancer. J Surg Oncol. 2005;89:114–21. doi: 10.1002/jso.20185. [DOI] [PubMed] [Google Scholar]
- 25.Mazzaferri EL. Management of low-risk differentiated thyroid cancer. Endocr Pract. 2007;13:498–512. doi: 10.4158/EP.13.5.498. [DOI] [PubMed] [Google Scholar]
- 26.Molinaro E, Pieruzzi L, Viola D. Radioiodine post-surgical remnant ablation in patients with differentiated thyroid cancer: news from the last 10 years. J Endocrinol Invest. 2012;35:16–20. [PubMed] [Google Scholar]
- 27.Frasoldati A, Pesenti M, Gallo M, Caroggio A, Salvo D, Valcavi R. Diagnosis of neck recurrences in patients with differentiated thyroid carcinoma. Cancer. 2003;97:90–6. doi: 10.1002/cncr.11031. [DOI] [PubMed] [Google Scholar]
- 28.Pacini F, Molinaro E, Castagna MG, Agate L, Elisei R, Ceccarelli C, Lippi F, Taddei D, Grasso L, Pinchera A. Recombinant human thyrotropin-stimulated serum thyroglobulin combined with neck ultrasonography has the highest sensitivity in monitoring differentiated thyroid carcinoma. J Clin Endocrinol Metab. 2003;88:3668–73. doi: 10.1210/jc.2002-021925. [DOI] [PubMed] [Google Scholar]
- 29.Whitley RJ, Ain KB. Thyroglobulin: a specific serum marker for the management of thyroid carcinoma. Clin Lab Med. 2004;24:29–47. doi: 10.1016/j.cll.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 30.Spencer CA. Challenges of serum thyroglobulin (Tg) measurement in the presence of Tg autoantibodies. J Clin Endocrinol Metab. 2004;89:3702–4. doi: 10.1210/jc.2004-0986. [DOI] [PubMed] [Google Scholar]
- 31.Ito Y, Higashiyama T, Takamura Y, Miya A, Kobayashi K, Matsuzuka F, Kuma K, Miyauchi A. Risk factors for recurrence to the lymph node in papillary thyroid carcinoma patients without preoperatively detectable lateral node metastasis: validity of prophylactic modified radical neck dissection. World J Surg. 2007;31:2085–91. doi: 10.1007/s00268-007-9224-y. [DOI] [PubMed] [Google Scholar]
- 32.Hsu YT, Gu F, Huang YW, Liu J, Ruan J, Huang RL, Wang CM, Chen CL, Jadhav RR, Lai HC, Mutch DG, Goodfellow PJ, Thompson IM, Kirma NB, Huang TH. Promoter hypomethylation of EpCAM-regulated bone morphogenetic protein gene family in recurrent endometrial cancer. Clin Cancer Res. 2013;19:6272–85. doi: 10.1158/1078-0432.CCR-13-1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stott-Miller M, Zhao S, Wright JL, Kolb S, Bibikova M, Klotzle B, Ostrander EA, Fan JB, Feng Z, Stanford JL. Validation study of genes with hypermethylated promoter regions associated with prostate cancer recurrence. Cancer Epidemiol Biomarkers Prev. 2014;23:1331–9. doi: 10.1158/1055-9965.EPI-13-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Su SF, de Castro Abreu AL, Chihara Y, Tsai Y, Andreu-Vieyra C, Daneshmand S, Skinner EC, Jones PA, Siegmund KD, Liang G. A panel of three markers hyper- and hypomethylated in urine sediments accurately predicts bladdercancer recurrence. Clin Cancer Res. 2014;20:1978–89. doi: 10.1158/1078-0432.CCR-13-2637. [DOI] [PubMed] [Google Scholar]
- 35.Mancikova V, Buj R, Castelblanco E, Inglada-Pérez L, Diez A, de Cubas AA, Curras-Freixes M, Maravall FX, Mauricio D, Matias-Guiu X, Puig-Domingo M, Capel I, Bella MR, Lerma E, Castella E, Reverter JL, Peinado MÁ, Jorda M, Robledo M. DNA methylation profiling of well-differentiated thyroid cancer uncovers markers of recurrencefree survival. Int J Cancer. 2014;135:598–610. doi: 10.1002/ijc.28703. [DOI] [PubMed] [Google Scholar]
- 36.Czarnecka K, Pastuszak-Lewandoska D, Migdalska-Sek M, Nawrot E, Brzezinski J, Dedecjus M, Pomorski L, Brzezianska E. Aberrant methylation as a main mechanism of TSGs silencing in PTC. Front Biosci (Elite Ed) 2011;3:137–57. doi: 10.2741/e228. [DOI] [PubMed] [Google Scholar]
- 37.Marsh S. Pyrosequencing applications. Methods Mol Biol. 2007;373:15–24. doi: 10.1385/1-59745-377-3:15. [DOI] [PubMed] [Google Scholar]
- 38.Huang G, Krocker JD, Kirk JL, Merwat SN, Ju H, Soloway RD, Wieck LR, Li A, Okorodudu AO, Petersen JR, Abdulla NE, Duchini A, Cicalese L, Rastellini C, Hu PC, Dong J. Evaluation of INK4A promoter methylation using pyrosequencing and circulating cell-free DNA from patients with hepatocellular carcinoma. Clin Chem Lab Med. 2014;52:899–909. doi: 10.1515/cclm-2013-0885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Christofori G, Semb H. The role of the cell-adhesion molecule E-cadherin as a tumour-suppressor gene. Trends Biochem Sci. 1999;24:73–6. doi: 10.1016/s0968-0004(98)01343-7. [DOI] [PubMed] [Google Scholar]
- 40.Canel M, Serrels A, Frame MC, Brunton VG. E-cadherin-integrin crosstalk in cancer invasion and metastasis. J Cell Sci. 2013;126:393–401. doi: 10.1242/jcs.100115. [DOI] [PubMed] [Google Scholar]
- 41.Blyth K, Cameron ER, Neil JC. The RUNX genes: gain or loss of function in cancer. Nat Rev Cancer. 2005;5:376–87. doi: 10.1038/nrc1607. [DOI] [PubMed] [Google Scholar]
- 42.Ko HJ, Kim BY, Jung CH, Chun SW, Mok JO, Kim YJ, Park HK, Kim CH, Kim SJ, Byun DW, Suh KI, Yoo MH, Kang SG. DNA methylation of RUNX3 in papillary thyroid cancer. Korean J Intern Med. 2012;27:407–10. doi: 10.3904/kjim.2012.27.4.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


