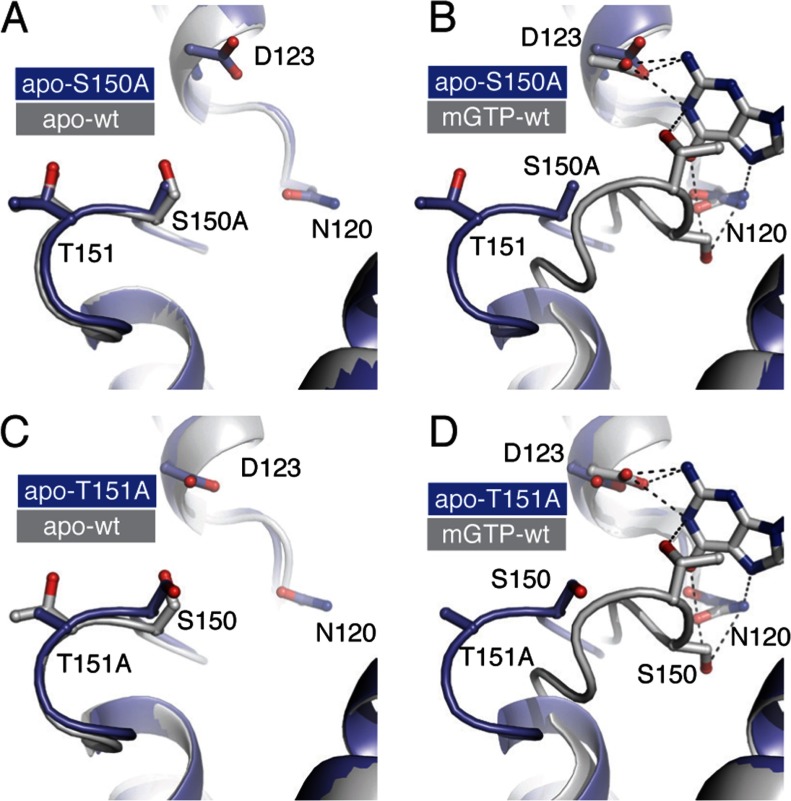
Figure 4. Structural validation and analysis of the S150A and T151A mutant proteins.
Structural overlay of the G5 loop of the S150A mutant with that of the (A) apo-wild type EcNFeoB (PDB ID 3HYR) and (B) mGTP bound wild type EcNFeoB (PDB ID 3HYT). The G5 loop of the S150A mutant is identical to the wild-type apo structure conformation and distinct from the nucleotide bound conformation. Bonds are shown as dotted line. (C,D) Structural overlay of the G5 loop of the T151A mutant with that of (C) apo-wild-type EcNFeoB and (D) mGTP-bound wild-type EcNFeoB. As with the S150A mutant structure, the T151A mutant structure is virtually identical in conformation to the wild-type apo EcNFeoB structure. The electrostatic or Van Der Waal interactions in the wild-type structure highlight the interactions between the T151 hydroxyl group with the N1 nitrogen of the nucleotide, and the hydrophobic interaction between the T151 methyl group and the nucleotide base, which are lost with the T151A mutation.