Abstract
Multiple novel simian adenoviruses have been isolated over the past years and their potential to cross the species barrier and infect the human population is an ever present threat. Here we describe the isolation and full genome sequencing of a novel simian adenovirus (SAdV) isolated from the urine of two independent, never co-housed, late stage simian immunodeficiency virus (SIV)-infected rhesus macaques. The viral genome sequences revealed a novel type with a unique genome length, GC content, E3 region and DNA polymerase amino acid sequence that is sufficiently distinct from all currently known human- or simian adenovirus species to warrant classifying these isolates as a novel species of simian adenovirus. This new species, termed Simian mastadenovirus D (SAdV-D), displays the standard genome organization for the genus Mastadenovirus containing only one copy of the fiber gene which sets it apart from the old world monkey adenovirus species HAdV-G, SAdV-B and SAdV-C.
Keywords: adenovirus, non-human primate, rhesus macaque, simian adenovirus, adenovirus species, virus genome analysis, virus phylogenomics
Introduction
The family Adenoviridae comprises a large family of non-enveloped double stranded DNA viruses. It can be subdivided into five genera which contain a plethora of viruses infecting a wide range of hosts including mammals, birds, fish, and amphibians (Wold and Horwitz, 2007). The genus Mastadenovirus includes 68 human types identified to date which divide into 7 known species (A–G) (Harrach et al., 2011). These types differ greatly in their tropism and pathogenicity, but they are generally known to infect the human respiratory-, urinary- or gastrointestinal tract or the conjunctiva causing a range of medical conditions including pneumonia, croup, bronchitis, keratoconjunctivitis, gastroenteritis and cystitis (Jones et al., 2007; Lewis et al., 2009; Louie et al., 2008; Saad et al., 1997; Treacy et al., 2010; Walsh et al., 2009; Wold and Horwitz, 2007; Wood, 1988). Adenoviral infections are usually innocuous, but can be serious in immunocompromised patients such as AIDS patients (Echavarria, 2008). Adenovirus types isolated from great apes are most closely related to human types and cluster together with them within the human adenovirus (HAdV) species A–F (Roy et al., 2009). In contrast, simian adenoviruses (SAdV) of old-world monkey (OWM) origin form separate species (A–C). Although adenoviruses are generally considered to be species specific, zoonotic infections have been reported (Gillespie et al., 2008; Wolfe et al., 2007). The recently described human type 52 (HAdV-52), the founding and so far only human type of the Human mastadenovirus species G (HAdV-G), was isolated from the stool of five different patients during an outbreak of gastroenteritis in Los Angeles county (Jones et al., 2007). Interestingly, although considered a human species, HAdV-G also includes multiple types isolated from OWM and no further types of human or great ape origin (Chiu et al., 2013; Roy et al., 2012; Wevers et al., 2011). HAdV-52 is thus likely of monkey origin and could therefore represent a documented case of an OWM simian adenovirus crossing the species barrier to cause disease in humans.
Besides being important human and animal pathogens, adenoviruses are also widely used as vectors in vaccine development and gene therapy. Human adenovirus 5 (HAdV-5) is by far the most widely used vector but it showed poor efficacy in HIV-1 clinical trials (Buchbinder et al., 2008; McElrath et al., 2008) presumably due to the fact that this type is widespread in the human population and preexisting humoral immune responses against HAdV-5 impair the immunogenicity of HAdV-5-vectored vaccines (Casimiro et al., 2003; Catanzaro et al., 2006; McElrath et al., 2008; Priddy et al., 2008). To bypass this problem animal adenoviruses are being developed as alternative to human vaccine vectors. Although simian adenoviruses of great ape origin are preferred because they belong to the same species as their human counterpart without showing significant seroprevalence in the human population (Dicks et al., 2012) adenoviruses of OWM origin could also conceivably be considered since they have shown the capability to naturally cross the species barrier and infect human individuals.
Here we describe the isolation and full genome analysis of a novel simian adenovirus type of rhesus macaque origin. Full genome analysis of our isolate using NextGen sequencing revealed a genome sequence substantially different from all previously described human and simian types and species. Therefore, we classify our novel simian adenovirus type as a novel species of simian adenoviruses termed Simian mastadenovirus D (SAdV-D).
Results
Isolation of simian adenovirus 26296 and 23336
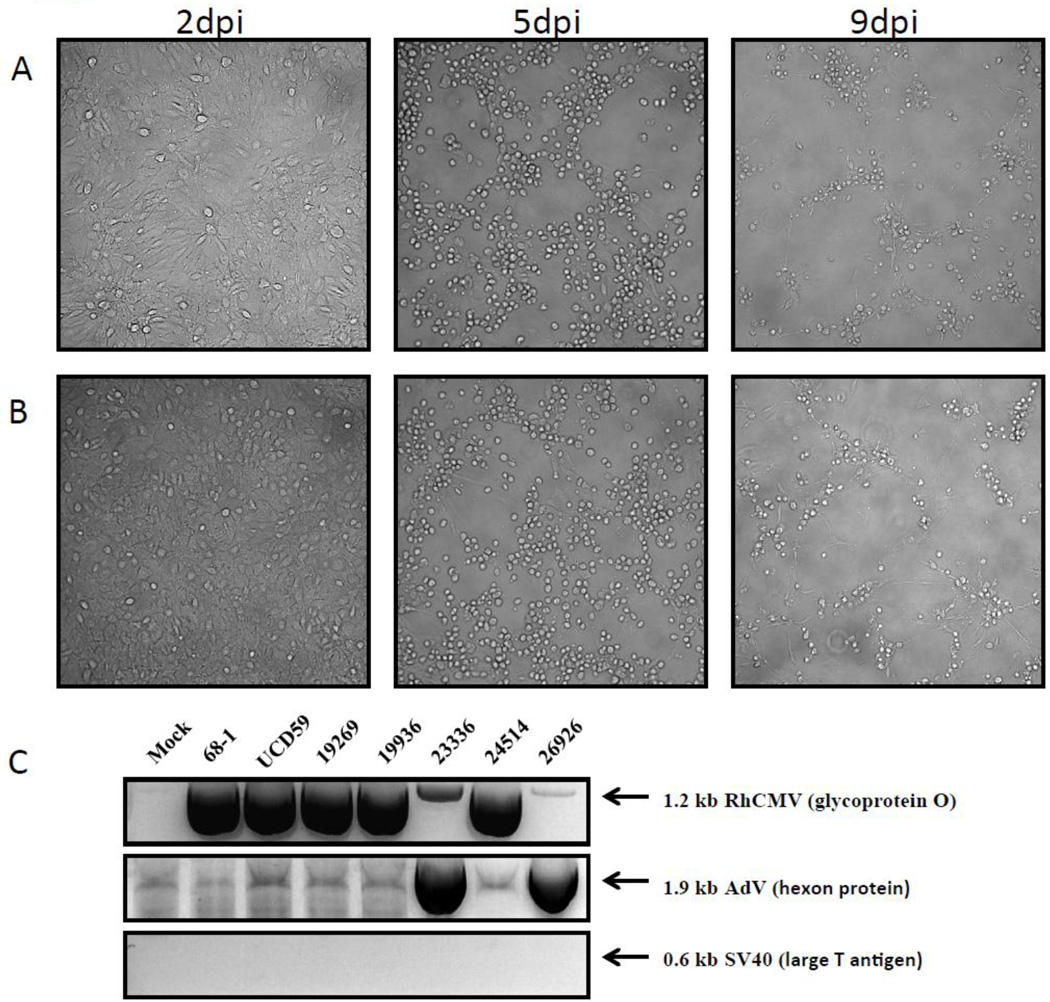
Rhesus macaques (RM) suffering from AIDS like symptoms demonstrate an SIV-associated expansion of the enteric virome including the increased appearance of multiple adenoviruses in the gastrointestinal tract (Handley et al., 2012). To determine whether SIV-infected RM secreted novel viruses in their urine we collected urine from several late stage SIVmac239-infected RM and spinoculated primary rhesus fibroblasts with the fresh samples. The cells were monitored in regular intervals and most samples developed owl eyed plaques within 1–2 weeks of infection indicative of cytomegalovirus (CMV) (data no shown). This observation was consistent with close to 100% of the animals in our cohort being naturally CMV+. However, one viral isolate showed a different phenotype of infection (Fig.1A) with infected cells resembling human foreskin fibroblasts infected with HAdV-5 (Rothmann et al., 1998). In a second experiment, samples were frozen prior to spinoculation, a process that should reduce live CMV contamination, and a second viral isolate with similar phenotype was obtained (Fig. 1B).
Fig.1. Isolation if simian adenovirus from the urine of late stage infected rhesus macaques.
Primary rhesus fibroblasts were infected with virus isolated from the urine of rhesus macaque 26296 (A) or rhesus macaque 23336 (B) at an MOI of 3 and phase contrast pictures were taken 2dpi (left panel), 5dpi (middle panel) and 9dpi (right panel). (C) Total DNA was isolated from primary rhesus fibroblast infected with virus isolated form rhesus macaques 19262, 19936, 23336, 24514 and 26296 after the infection reached full CPE and the presence of either RhCMV (gO), adenovirus (hexon) or SV40 (large T antigen) was detected by PCR using specific primers. The RhCMV laboratory strain 68-1 and the primary RhCMV isolate UCD59 were included as positive controls for RhCMV.
To examine whether these newly isolated viruses were adenoviruses, we isolated DNA from primary rhesus fibroblasts inoculated with viral isolates from the urine of late stage-SIV infected rhesus macaques (19262, 19936, 23336, 24514 and 26296) or RhCMV strains 68-1 and UCD59 as positive RhCMV controls and performed diagnostic PCRs with primers for RhCMV gO, for the adenovirus hexon protein (Roy et al., 2009) or, as negative control, the large T antigen of SV40 (Bergsagel et al., 1992). As shown in Fig.1C, all virus isolates or controls tested were negative for SV40, whereas all putative CMV isolates were positive for gO-specific primers. RhCMV gO-specific bands were also observed in the putative adenovirus-containing isolates, albeit very faint. In addition, both putative adenoviruses were the only isolates that tested positive for the hexon gene consistent with both isolates containing adenovirus sequences. We termed the two isolates SAdV-26296 and SAdV-23336. Sequence analysis of the gO-specific PCR fragments perfectly matched RhCMV gO in both cases consistent with RhCMV contamination in both SAdV-isolates. In addition, both SAdV contained a hexon gene fragment that was identical in sequence.
Genome sequencing and analysis of SAdV-23336
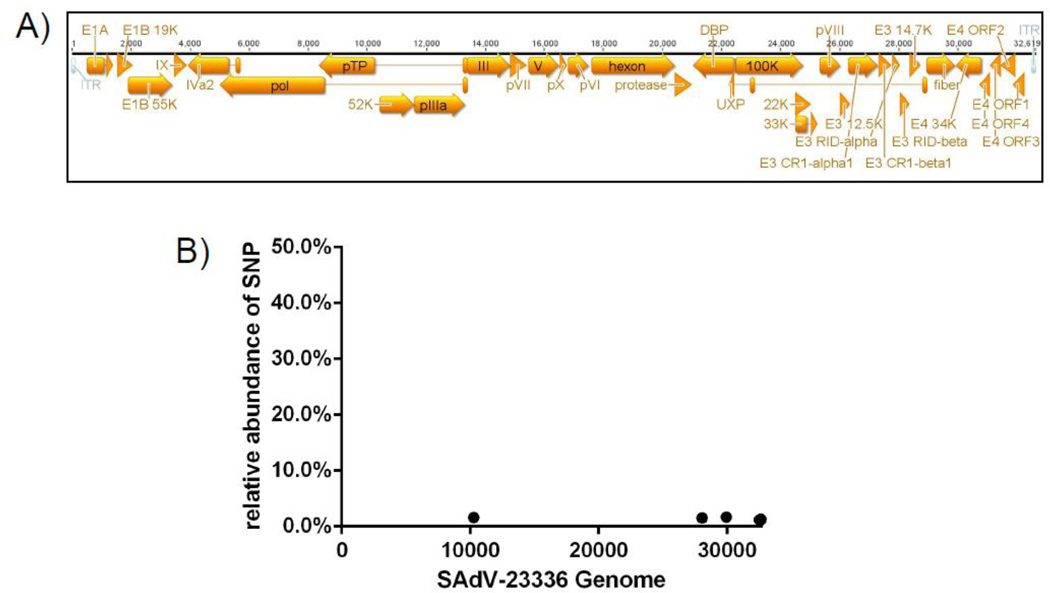
Although the sequences of the hexon gene fragments were identical between the two isolated viruses they differed significantly from all hexon sequences present in GenBank. To further determine the entire genome sequence we performed a full genome analysis using next generation sequencing. Since the hexon gene sequences were identical between the isolates we selected only one isolate (SAdV-23336) for further analysis. This isolate showed significantly less RhCMV contamination (Fig.1C). Since herpesviruses are opportunistic pathogens and as such reactivate in the immunocompromised host, the difference in RhCMV genome copies detected in our two isolates might reflect individual difference in the amount of CMV shedd by the late stage SIV infected animals. Viral DNA was isolated from the supernatant of infected primary rhesus fibroblasts and the isolated DNA was sequenced on an Illumina MiSeq next generation sequencer. Using Geneious 6.1.7 we assembled the full viral genome of 32.661 bp with a 1150 fold coverage. 54.2% of our reads were specific to SAdV-23336 whereas none of the reads could be aligned to RhCMV, indicating that the initial CMV contamination was lost during passaging in tissue culture. The resulting adenovirus genome is representative of the genus Mastadenovirus with all viral genes generally found in human- or simian adenoviruses present with the exceptions of E4orf6/7 (Fig.2A), a viral transcription regulatory protein (Huang and Hearing, 1989; Marton et al., 1990; Nevins, 1992; Obert et al., 1994; Raychaudhuri et al., 1990) involved in cell transformation (Yamano et al., 1999). Closer examination of the deep sequencing data revealed only eight single nucleotide polymorphisms (SNPs) with a frequency above 1% with the highest being only 1.7 % of all reads in that position (Fig.2B). Since such low frequency SNPs are most likely sequencing errors rather than true polymorphisms (Loman et al., 2012) these results indicate a very homogenous viral population rather than a mixture of different adenovirus strains.
Fig.2. Full genome sequence and ORF annotation of SAdV-23336.
Viral DNA was isolated from the supernatant of SAdV-23336 infected rhesus fibroblasts. Next-generation sequencing was performed using an Illumina MiSeq sequencer and the full genome was assembled using Geneious 6.1.7. A) Graphical depiction of the annotated full genome sequence of SAdV-23336. B) Single nucleotide polymorphisms (SNPs) with an incident rate of above 1% are shown across the entire genome. Only 8 SNPs were found at low frequency none of them exceeding 1.7%.
Phylogenetic analysis of SAdV-23336
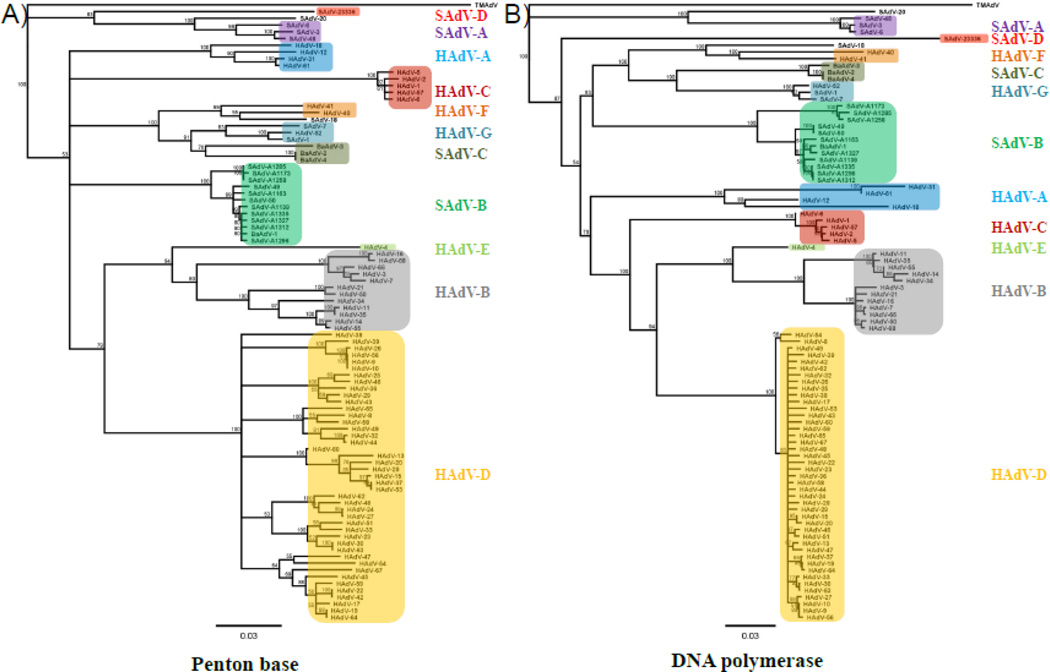
To establish the evolutionary relationship of this new isolate to other adenoviruses we compared the amino acid sequence of the penton base and of the DNA polymerase to all 68 known human types as well as all isolates of rhesus macaque (SAdV-3, SAdV-6, SAdV-7, SAdV-A1139, SAdV-A1163, SAdV-A1173, SAdV-A1258, SAdV-A1285, SAdV-A1296, SAdV-A1312, SAdV-A1327 and SAdV-A1335), cynomolgus macaque (SAdV-1, SAdV-48, SAdV-49 and SAdV-50), vervet monkey (SAdV-18 and SAdV-20) and olive baboon (BaAdV-1, BaAdV-2, BaAdV-3 and BaAdV-4) origin published to date. To begin our phylogenetic analysis we imported the predicted amino acid sequences for the penton base and the DNA polymerase of the aforementioned types from GenBank into Geneious 6.1.7. and generated full amino acid sequence alignments using the Geneious sequence alignment algorithm. Subsequently, amino acid distance-based unrooted neighbor-joining trees were generated based on the alignments using Geneious Tree Builder applying the neighbo-joining method. Using titi monkey adenovirus (TMAdV) isolated from a NWM as an outgroup, bootstrap analysis with 100 resamplings from the aligned sequences was applied to test the validity of the tree topology followed by distance matrix calculations and calculation of the most probable consensus tree with a support threshold of 50%. The resulting phylogenetic trees based on all penton base (Fig.3A) or DNA polymerase (Fig.3B) sequences resulted in each type grouping into the expected species thus supporting our analysis method. However, when the penton base or the DNA polymerase of our new isolate was included in the phylogenetic analysis, none of the known adenovirus species showed a close evolutionary relationship due to the low homology of these proteins between our new isolate and all previously described adenovirus types.
Fig.3. SAdV-23336 forms a distinct phenotypic cluster when compared to known species of human-or simian adenoviruses.
All 68 HAdV types known to date as well as all SAdV isolates from rhesus macaques, cynomolgus macaques, vervet monkeys or baboons were used to create phylogenetic trees. The amino acid sequences of either the penton base (A) or the DNA polymerase (B) were aligned using Geneious 6.1.7 and phylogenetic trees were generated using the neighbor-joining method. TMAdV was used as an outgroup.
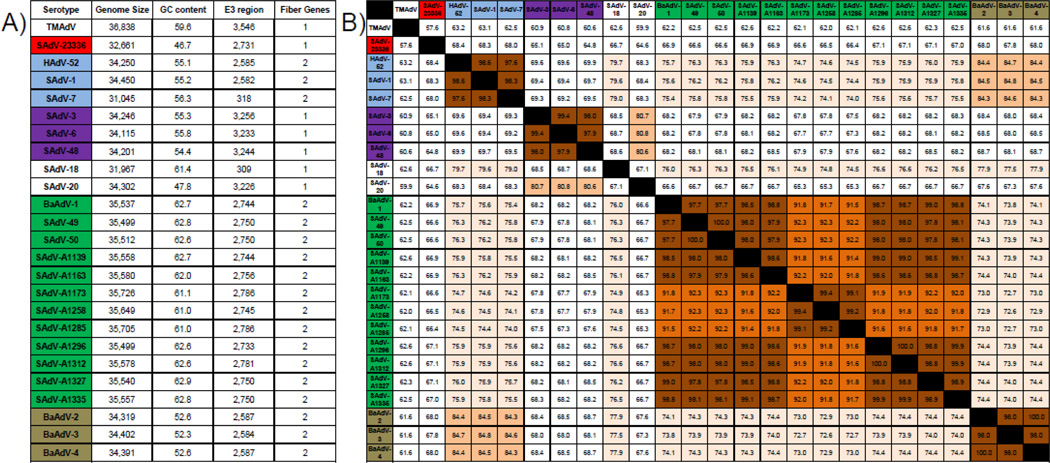
Although all human and simian members of the genus Mastadenovirus have the same overall genome organization, they differ in the number of encoded fiber genes. Among human adenoviruses, the species A–E encode one fiber gene, whereas the species HAdV-F and HAdV-G contain two different fiber genes. Within the OWM-derived simian adenoviruses, only types within the species SAdV-A and the non-classified types SAdV-18 and SAdV-20 encode one fiber gene, whereas all members of the species SAdV-B and SAdV-C as well as all human and simian types grouped together in the Human mastadenovirus species HAdV-G encode two different fiber genes (Fig.4A). Similar to SAdV-A only a single copy of the fiber gene was encoded by the new isolate SAdV-23336. However, the predicted amino acid sequence for the fiber protein was considerably shorter compared to all types grouped in SAdV-A and exhibited very low overall identity (<30%). In addition, our isolate displays a unique GC content (46.7%) that is notably lower than the GC content in any other classified SAdV type. Since the overall GC content is usually highly conserved between members of the same species this indicates that SAdV-23336 is distinct from all known types and does not fall into any established species. Similarly, the length of the E3 region encoded in the viral genome, which can be used to insert foreign antigens in vector development, as well as the full genome length differ greatly between species, but not between types within a species. The E3 region of SAdV-23336 contains 2731 bp ranging from the start codon of the predicted E3 12.5kDa protein to the stop codon of the predicted E3 14.7 kDa protein. In comparison to the other examined SAdV species, this length resembles the E3 region found in species SAdV-B, but it differs substantially from the E3 region found in HAdV-G, SAdV-A and SAdV-C. Furthermore, the SAdV-23336 genome length of 32,661 bp is considerably shorter than the genomes of representative members of all other established SAdV species. The types SAdV-7 and SAdV-18 are outliers of this general rule since their E3 region as well as subsequently their full length genome are significantly shorter than those of other types of human or simian adenoviruses, but it has been noted before that this is likely due to a truncation of the viral genome (Roy et al., 2011). Lastly, the comparison of the conserved DNA polymerase genes across all simian adenovirus species (Fig.4B) revealed that different types of the same species are more than 90% identical, whereas types that belong to different species are less than 80% identical. SAdV-23336 does not share more than 70% identity to any other SAdV type. Thus, the new isolate described here is significantly different from any previously defined species of simian adenoviruses, indicating that SAdV-23336 might represent a new species within the genus Mastadenovirus for which we propose the name Simian mastadenovirus D (SAdV-D).
Fig.4. SAdV-23336 differs substantially from known simian adenovirus types.
A) Different SAdV species vary in their GC content, the length and coding content of their E3 region as well as their overall genome size and in the number of fiber genes they encode. A combination of all these characteristics can be used to classify types into the species they belong to. B) A further characteristic is the level of conservation between the DNA polymerase across species and types. The total level of identity shown as per cent (%) identity between the DNA polymerase amino acid sequences of the indicated SAdV types was determined using the same analysis as for the phylogenetic tree shown in figure 3A. The different shades of orange indicate different levels of identity (≥70%, ≥80%, ≥90% and ≥95%). Types highlighted in light blue (HAdV-G), purple (SAdV-A), green (SAdV-B) and brown (SAdV-C) indicate the different analyzed species. Our novel isolate is shown in red (SAdV-D). SAdV-18 and SAdV-20 have not been classified yet, but given that these two types do not show any significant similarity in length, number of fiber genes, GC content, and length of their E3-region to any other human or simian type or to each other, it is possible that they too represent independent simian adenovirus species. The NWM simian adenovirus TMAdV was used for comparison.
Discussion
We report here on the isolation and genomic characterization of a novel simian adenovirus from secretions of late stage SIV-infected RM. The genomic sequence of this virus differs significantly from any known human- or simian adenovirus suggesting that we isolated a novel species. Interestingly, the two monkeys used to isolate the viruses were never co-housed suggesting that these SAdV are widespread in our colony. The potential of this virus to infect humans is currently not known, but the recent identification of several novel human and simian adenoviruses and their phylogenetic analysis suggests that cross species infections among human and non-human primates might occur more frequently than previously assumed. The human adenovirus type 52 was isolated in Los Angeles county during an outbreak of gastroenteritis from stool specimen of human patients (Jones et al., 2007). This virus differed substantially from the previously known human species A–F and displayed a duplication of the fiber gene, a genotype that is rarely found in human adenoviruses. It became the founding member of a novel human adenovirus species termed HAdV-G, but even after years of further research, no additional human or great ape type grouping into the same species could be identified. In contrast, multiple simian types of OWM origin exhibited high similarity to HAdV-52, so that the majority of types clustering in the human species HAdV-G were actually isolates from rhesusand cynomolgus macaques, suggesting that HAdV-52 might have entered the human population via zoonotic infection from monkeys (Chiu et al., 2013; Maluquer de Motes et al., 2011; Roy et al., 2012; Roy et al., 2009; Wevers et al., 2011). In another example, a respiratory infection that occurred in a researcher at the California National Primate Research Center (CNPRC) during an outbreak of pneumonia and hepatitis in a colony of titi monkeys could be linked to a novel adenovirus (TMAdV) (Chen et al., 2011). This observation suggested that adenoviruses can leap from rather distantly related new world monkeys (NWM) to humans. Moreover, human to human transmission of TMAdV was observed in a close family member of the researcher originally infected (Chen et al., 2011). Taken together, these observations suggest that novel adenoviruses can be contracted from NHP and that our newly identified OWM AdV species might represent a risk for animal caretakers.
Sequence analysis and genomic comparisons suggests that our adenovirus isolate represents a novel type of simian adenovirus, although we did not perform any serology experiments to determine cross neutralization with other closely related types. The low sequence homology to any known human or simian adenoviruses further suggests that this isolate represents a novel species as well. Among the criteria used to define adenovirus species in the 2011 ICTV report, different species generally display a phylogenetic distance of >5–15%, based primarily on distance matrix analysis of the DNA polymerase amino acid sequence, a genome organization, characteristically in the E3 region, that is unique to members of a certain species and GC content that varies greatly between different species (Harrach et al., 2011). SAdV-23336 shows no similarity in GC content to any of the established SAdV species and only some similarity to SAdV-B when examining the length of the E3 region and to SAdV-A when comparing the number of encoded fiber genes. In addition, the viral genome is unusually short compared to all other SAdV of OWM origin and lacks the control protein E4orf6/7 generally found in primate adenoviruses. Since our isolate shows less than 70% identity (more than 30% distance) to any other published amino acid sequence of the DNA polymerase we propose that this adenovirus represents a novel species and type which will likely be confirmed by future serological analysis. Given the substantial difference between our novel isolate and all other human and simian species described to date we propose a new species termed Simian mastadenovirus D (SAdV-D). It will be interesting to examine how widespread this species is disseminated among our colony of Indian-origin RM and potentially among humans in contact with these animals.
Materials and Methods
Cells
Primary rhesus fibroblasts were maintained in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum, 100 U/ml penicillin, and 100 µg/ml streptomycin and were grown at 37°C in humidified air with 5% CO2.
Isolation of the novel simian adenoviruses
Urine was obtained from SIVmac239-infected rhesus macaques either through cystocentesis or during necropsy. The urine was first centrifuged at 2000 g for 10 minutes at 4°C to pellet all cell debris and contaminants, and then filtered through a 0.45 µm filter (Millipore) to clear the urine from any bacterial or fungal contamination. Subsequently, primary rhesus fibroblasts seeded out in 6 well plates were spinoculated by adding 500 µl –1000 µl of cleared urine per well and centrifugation at 700 g for 30 minutes at 25°C. 2hpi the cells were washed and new DMEM + 10% fetal bovine serum and penicillin/streptomycin was added. The cells were kept in culture for up to 1 month or until signs of viral infection were noticed in which case the cells were harvested at full cytopathic effect (CPE) and the virus was purified and analyzed.
SAdV purification and stock generation
SAdV was grown on primary rhesus fibroblasts until full CPE. Cells and supernatant were harvested, pooled and frozen to release residual virus from the cells. The cellular debris was removed in two steps, first by centrifugation at 2000 g for 10 minutes at 4°C and subsequently by centrifugation at 7500 g for 15 minutes. The clarified medium was layered over a sorbitol cushion (20% D-sorbitol, 50 mM Tris [pH 7.4], 1 mM MgCl2), and virus was spun down by centrifugation at 64,000 g for 1 hour at 4°C in a Beckman SW28 rotor. The virus pellet was resuspended in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum, 100 U/ml penicillin, and 100 µg/ml streptomycin, aliquoted, snap frozen in liquid nitrogen and stored at −80°C until further use. Viral titers were determined by limited dilution assay on primary rhesus fibroblast and CPE was determined in infected wells after 14 days.
Identification of the novel isolates using PCR
Primary rhesus fibroblasts were infected with viral isolates at an multiplicity of infection (MOI) of 3 and the cells were harvested at full CPE. DNA was isolated using a DNeasy Blood & Tissue Kit (Qiagen). To test for the presence of RhCMV, Adenoviruses and SV40 the following primers were used: RhCMV gO (Sense: 5’-TGCGCATTGTACTTCGCAT-3’; Antisense: 5’-GTATTTATTCAACGAGACGTA-3’), a conserved region in the adenovirus hexon protein (Sense: 5’-CAGGACGCCTCGGAGTACCTGAG-3’; Antisense: 5’- TTGGCAGGAATGGGGTACAGCATGTT-3’) (Roy et al., 2009), the large T antigen of SV40 (Sense: 5’-GCATGACTCAAAAAACTTAGCAATTCTG-3’; Antisense: 5’-CTTTGGAGGCTTCTGGGATGCAACT-3’) (Bergsagel et al., 1992). PCR was performed using AccuPrime™ Taq DNA Polymerase High Fidelity (Life Technologies) following the manufacturer’s instructions. The amplified DNA was purified on a 1% agarose gel and the isolated DNA was sent to Eurofins MWG Operon (Huntsville, AL, USA) for sequencing.
Next generation sequencing of viral DNA
To purify viral DNA we modified a protocol previously used for the extraction of viral DNA from HAdV-40 (Mautner, 2007). Primary Rhesus fibroblasts were infected with SAdV at an MOI of 0.1 and the supernatant was harvested at full CPE. To remove cellular contaminants, residual cells and cell debris were removed by centrifugation initially at 2000 g for 10 minutes at 4°C and subsequently at 7500 g for 15 minutes. To generate a viral pellet, the clarified medium was layered over a sorbitol cushion (20% D-sorbitol, 50 mM Tris [pH 7.4], 1 mM MgCl2), and virus was concentrated by centrifugation at 64,000 g for 1 hour at 4°C in a Beckman SW28 rotor. The virus pellet was resuspended in 500µl 10.1 TE Buffer (10mM Tris, pH 8.0; 0.1mM EDTA, pH 8.0) and 500 µl 2× Lysis Buffer (20mM Tris-Cl, pH 8.0; 50mM EDTA, pH8.0; 200mM NaCl; 1.2% w/v SDS) were added to lyse the viral virions. Finally, 250µg Proteinase K were added and the solution was incubated for 2h at 37°C. The viral DNA was phenol/chloroform extracted twice and precipitated with absolute ethanol at −80°C overnight. The DNA was spun down at 16000 g for 20 minutes at 4°C and washed once with 70% ethanol. Finally the DNA was resuspended in water and the DNA concentration was determined using a ND-1000 Spectrophotometer (NanoDrop Technologies, Inc.).
Generation of Next Generation Sequencing libraries
The DNA was fragmented using an S2 Sonicator (Covaris) and was then converted to libraries using the standard TruSeq protocol (Illumina). The ends of the fragmented DNA were enzymatically blunted, then a single terminal “A” nucleotide was added to assist ligation. Adaptors containing flow cell-binding sequences, indices, and primer binding sites were then ligated to the fragments. The resulting DNA was amplified using polymerase chain reaction with a limited number of cycles. Following removal of adaptor dimers, the library was examined on the Bioanalyzer (Agilent) and the concentration was determined using real time PCR and SYBR Green fluorescence.
Next Generation sequencing
Next generation sequencing was performed using a MiSeq Next-Generation Sequencing System (Illumina). The generated libraries were loaded into a MiSeq reagent cartridge at a concentration of 9 pM and single read sequencing was performed for 300 cycles with 6 additional cycles of index reads. The resulting data was imported into Geneious 6.1.7. and the sequencing reads were trimmed of all regions exceeding the error probability limit of 0.1% to minimize sequencing errors. Subsequently, the genome of our viral isolate was de novo assembled using the processed sequencing data.
Generation of phylogenetic trees and statistical analysis
Relevant predicted amino acid sequences of the adenoviral proteins of interest (penton base and DNA polymerase) were imported from GenBank into Geneious 6.1.7. and amino acid sequences alignments were generated using Geneious Alignment. Phylogenetic analysis to generate amino acid distancebased unrooted neighbor-joining trees (Saitou and Nei, 1987) was performed based on these alignments using Geneious Tree Builder applying the neighbor-joining method. The validity of the tree topology obtained was tested by using bootstrap analysis (Felsenstein, 1985) with 100 resamplings from the aligned sequences, followed by distance matrix calculations and calculation of the most probable consensus tree with a support threshold of 50%. In all shown phylogenetic trees, the NWM adenovirus TMAdV was used as an outgroup.
Viral sequences utilized in this study
The full genome sequence of the newly isolated simian adenovirus 23336 (SAdV-23336) was submitted to GenBank (KM190146). The following adenovirus sequences published in GenBank were used in this study: BaAdV-1 (KC693021), BaAdV-2 (KC693022), BaAdV-3 (KC693023), BaAdV-4 (KC693024), HAdV-1 (AC_000017), HAdV-2 (AC_000007), HAdV-3 (AY599834), HAdV-4 (AY458656), HAdV-5 (AC_000008), HAdV-6 (HQ413315), HAdV-7 (AC_000018), HAdV-8 (AB448769), HAdV-9 (AJ854486), HAdV-10 (JN226746), HAdV-11 (AF532578), HAdV-12 (X73487), HAdV-13 (JN226747), HAdV-14 (AY803294), HAdV-15 (JN226748), HAdV-16 (JN860680), HAdV-17 (HQ910407), HAdV-18 (GU191019), HAdV-19 (AB448774), HAdV-20 (JN226749), HAdV-21 (AY601633), HAdV-22 (FJ404771), HAdV-23 (JN226750), HAdV-24 (JN226751), HAdV-25 (JN226752), HAdV-26 (EF153474), HAdV-27 (JN226753), HAdV-28 (FJ824826), HAdV-29 (JN226754), HAdV-30 (JN226755), HAdV-31 (AM749299), HAdV-32 (JN226756), HAdV-33 (JN226758), HAdV-34 (AY737797), HAdV-35 (AY271307), HAdV-36 (GQ384080), HAdV-37 (AB448777), HAdV-38 (JN226759), HAdV-39 (JN226760), HAdV-40 (NC_001454), HAdV-41 (DQ315364), HAdV-42 (JN226761), HAdV-43 (KC529648), HAdV-44 (JN226763), HAdV-45 (JN226764), HAdV-46 (AY875648), HAdV-47 (JN226757), HAdV-48 (EF153473), HAdV-49 (DQ393829), HAdV-50 (AY737798), HAdV-51 (JN226765), HAdV-52 (DQ923122), HAdV-53 (AB605244), HAdV-54 (NC_012959), HAdV-55 (JX123028), HAdV-56 (HM770721), HAdV-57 (HQ003817), HAdV-58 (HQ883276), HAdV-59 (JF799911), HAdV-60 (HQ007053), HAdV-61 (JF964962), HAdV-62 (JN162671), HAdV-63 (JN935766), HAdV-64 (JQ326207), HAdV-65 (AP012285), HAdV-66 (JN860676), HAdV-67 (AP012302), HAdV-68 (JN860678), SAdV-1 (NC_006879), SAdV-3 (NC_006144), SAdV-6 (JQ776547), SAdV-7 (DQ792570), SAdV-18 (FJ025931), SAdV-20 (NC_020485), SAdV-48 (HQ241818), SAdV-49 (NC_015225), SAdV-50 (HQ241820), SAdV-A1139 (JN880448), SAdV-A1163 (JN880449), SAdV-A1173 (JN880450), SAdV-A1258 (JN880451), SAdV-A1285 (JN880452), SAdV-A1296 (JN880453), SAdV-A1312 (JN880454), SAdV-A1327 (JN880455), SAdV-A1335 (JN880456), TMAdV (NC_020487).
Acknowledgements
This project was supported by the National Center for Research Resources and the Office of Research Infrastructure Programs (ORIP) of the National Institutes of Health through grant P51OD011092. We are grateful to Dr. Robert Searles and the OHSU Massively Parallel Sequencing Shared Resource (MPSSR) for generating the NGS libraries and to Yibing Jia and the Molecular and Cellular Biology Core (MCB Core) at the Oregon National Primate Research Center (ONPRC) for analyzing the samples on their MiSeq next generation sequencer.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bergsagel DJ, Finegold MJ, Butel JS, Kupsky WJ, Garcea RL. DNA sequences similar to those of simian virus 40 in ependymomas and choroid plexus tumors of childhood. N Engl J Med. 1992;326:988–993. doi: 10.1056/NEJM199204093261504. [DOI] [PubMed] [Google Scholar]
- Buchbinder SP, Mehrotra DV, Duerr A, Fitzgerald DW, Mogg R, Li D, Gilbert PB, Lama JR, Marmor M, Del Rio C, McElrath MJ, Casimiro DR, Gottesdiener KM, Chodakewitz JA, Corey L, Robertson MN. Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step Study): a double-blind, randomised, placebo-controlled, test-of- concept trial. Lancet. 2008;372:1881–1893. doi: 10.1016/S0140-6736(08)61591-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casimiro DR, Chen L, Fu TM, Evans RK, Caulfield MJ, Davies ME, Tang A, Chen M, Huang L, Harris V, Freed DC, Wilson KA, Dubey S, Zhu DM, Nawrocki D, Mach H, Troutman R, Isopi L, Williams D, Hurni W, Xu Z, Smith JG, Wang S, Liu X, Guan L, Long R, Trigona W, Heidecker GJ, Perry HC, Persaud N, Toner TJ, Su Q, Liang X, Youil R, Chastain M, Bett AJ, Volkin DB, Emini EA, Shiver JW. Comparative immunogenicity in rhesus monkeys of DNA plasmid, recombinant vaccinia virus, and replication-defective adenovirus vectors expressing a human immunodeficiency virus type 1 gag gene. J Virol. 2003;77:6305–6313. doi: 10.1128/JVI.77.11.6305-6313.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catanzaro AT, Koup RA, Roederer M, Bailer RT, Enama ME, Moodie Z, Gu L, Martin JE, Novik L, Chakrabarti BK, Butman BT, Gall JG, King CR, Andrews CA, Sheets R, Gomez PL, Mascola JR, Nabel GJ, Graham BS. Phase 1 safety and immunogenicity evaluation of a multiclade HIV-1 candidate vaccine delivered by a replication-defective recombinant adenovirus vector. J Infect Dis. 2006;194:1638–1649. doi: 10.1086/509258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EC, Yagi S, Kelly KR, Mendoza SP, Tarara RP, Canfield DR, Maninger N, Rosenthal A, Spinner A, Bales KL, Schnurr DP, Lerche NW, Chiu CY. Cross-species transmission of a novel adenovirus associated with a fulminant pneumonia outbreak in a new world monkey colony. PLoS Pathog. 2011;7:e1002155. doi: 10.1371/journal.ppat.1002155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu CY, Yagi S, Lu X, Yu G, Chen EC, Liu M, Dick EJ, Jr, Carey KD, Erdman DD, Leland MM, Patterson JL. A novel adenovirus species associated with an acute respiratory outbreak in a baboon colony and evidence of coincident human infection. MBio. 2013;4:e00084. doi: 10.1128/mBio.00084-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dicks MD, Spencer AJ, Edwards NJ, Wadell G, Bojang K, Gilbert SC, Hill AV, Cottingham MG. A novel chimpanzee adenovirus vector with low human seroprevalence: improved systems for vector derivation and comparative immunogenicity. PLoS One. 2012;7:e40385. doi: 10.1371/journal.pone.0040385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echavarria M. Adenoviruses in immunocompromised hosts. Clin Microbiol Rev. 2008;21:704–715. doi: 10.1128/CMR.00052-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Gillespie TR, Nunn CL, Leendertz FH. Integrative approaches to the study of primate infectious disease: implications for biodiversity conservation and global health. Am J Phys Anthropol Suppl. 2008;47:53–69. doi: 10.1002/ajpa.20949. [DOI] [PubMed] [Google Scholar]
- Handley SA, Thackray LB, Zhao G, Presti R, Miller AD, Droit L, Abbink P, Maxfield LF, Kambal A, Duan E, Stanley K, Kramer J, Macri SC, Permar SR, Schmitz JE, Mansfield K, Brenchley JM, Veazey RS, Stappenbeck TS, Wang D, Barouch DH, Virgin HW. Pathogenic simian immunodeficiency virus infection is associated with expansion of the enteric virome. Cell. 2012;151:253–266. doi: 10.1016/j.cell.2012.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrach B, Benkö M, Both G, Brown M, Davis A, Echavarría M, Hess M, Jones M, Kajon A, Lahmkuhl H, Mautner V, Mittal S, Wadell G. Family Adenoviridae. In: King A, Adams M, Carstens E, Lefkowitz E, editors. Virus taxonomy: classification and nomenclature of viruses. Ninth report of the International Committee on Taxonomy of Viruses. San Diego, CA: Elsevier; 2011. pp. 95–111. [Google Scholar]
- Huang MM, Hearing P. The adenovirus early region 4 open reading frame 6/7 protein regulates the DNA binding activity of the cellular transcription factor, E2F, through a direct complex. Genes Dev. 1989;3:1699–1710. doi: 10.1101/gad.3.11.1699. [DOI] [PubMed] [Google Scholar]
- Jones MS, 2nd, Harrach B, Ganac RD, Gozum MM, Dela Cruz WP, Riedel B, Pan C, Delwart EL, Schnurr DP. New adenovirus species found in a patient presenting with gastroenteritis. J Virol. 2007;81:5978–5984. doi: 10.1128/JVI.02650-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis PF, Schmidt MA, Lu X, Erdman DD, Campbell M, Thomas A, Cieslak PR, Grenz LD, Tsaknardis L, Gleaves C, Kendall B, Gilbert D. A community-based outbreak of severe respiratory illness caused by human adenovirus serotype 14. J Infect Dis. 2009;199:1427–1434. doi: 10.1086/598521. [DOI] [PubMed] [Google Scholar]
- Loman NJ, Misra RV, Dallman TJ, Constantinidou C, Gharbia SE, Wain J, Pallen MJ. Performance comparison of benchtop high-throughput sequencing platforms. Nat Biotechnol. 2012;30:434–439. doi: 10.1038/nbt.2198. [DOI] [PubMed] [Google Scholar]
- Louie JK, Kajon AE, Holodniy M, Guardia-LaBar L, Lee B, Petru AM, Hacker JK, Schnurr DP. Severe pneumonia due to adenovirus serotype 14: a new respiratory threat? Clin Infect Dis. 2008;46:421–425. doi: 10.1086/525261. [DOI] [PubMed] [Google Scholar]
- Maluquer de Motes C, Hundesa A, Almeida FC, Bofill-Mas S, Girones R. Isolation of a novel monkey adenovirus reveals a new phylogenetic clade in the evolutionary history of simian adenoviruses. Virol J. 2011;8:125. doi: 10.1186/1743-422X-8-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marton MJ, Baim SB, Ornelles DA, Shenk T. The adenovirus E4 17-kilodalton protein complexes with the cellular transcription factor E2F, altering its DNA-binding properties and stimulating E1A-independent accumulation of E2 mRNA. J Virol. 1990;64:2345–2359. doi: 10.1128/jvi.64.5.2345-2359.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mautner V. Growth and purification of enteric adenovirus type 40. Methods Mol Med. 2007;130:145–156. doi: 10.1385/1-59745-166-5:145. [DOI] [PubMed] [Google Scholar]
- McElrath MJ, De Rosa SC, Moodie Z, Dubey S, Kierstead L, Janes H, Defawe OD, Carter DK, Hural J, Akondy R, Buchbinder SP, Robertson MN, Mehrotra DV, Self SG, Corey L, Shiver JW, Casimiro DR. HIV-1 vaccine-induced immunity in the test-of-concept Step Study: a case-cohort analysis. Lancet. 2008;372:1894–1905. doi: 10.1016/S0140-6736(08)61592-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevins JR. E2F: a link between the Rb tumor suppressor protein and viral oncoproteins. Science. 1992;258:424–429. doi: 10.1126/science.1411535. [DOI] [PubMed] [Google Scholar]
- Obert S, O'Connor RJ, Schmid S, Hearing P. The adenovirus E4-6/7 protein transactivates the E2 promoter by inducing dimerization of a heteromeric E2F complex. Mol Cell Biol. 1994;14:1333–1346. doi: 10.1128/mcb.14.2.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priddy FH, Brown D, Kublin J, Monahan K, Wright DP, Lalezari J, Santiago S, Marmor M, Lally M, Novak RM, Brown SJ, Kulkarni P, Dubey SA, Kierstead LS, Casimiro DR, Mogg R, DiNubile MJ, Shiver JW, Leavitt RY, Robertson MN, Mehrotra DV, Quirk E. Safety and immunogenicity of a replication-incompetent adenovirus type 5 HIV-1 clade B gag/pol/nef vaccine in healthy adults. Clin Infect Dis. 2008;46:1769–1781. doi: 10.1086/587993. [DOI] [PubMed] [Google Scholar]
- Raychaudhuri P, Bagchi S, Neill SD, Nevins JR. Activation of the E2F transcription factor in adenovirus-infected cells involves E1A-dependent stimulation of DNA-binding activity and induction of cooperative binding mediated by an E4 gene product. J Virol. 1990;64:2702–2710. doi: 10.1128/jvi.64.6.2702-2710.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothmann T, Hengstermann A, Whitaker NJ, Scheffner M, zur Hausen H. Replication of ONYX-015, a potential anticancer adenovirus, is independent of p53 status in tumor cells. J Virol. 1998;72:9470–9478. doi: 10.1128/jvi.72.12.9470-9478.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Clawson DS, Adam VS, Medina A, Wilson JM. Construction of gene transfer vectors based on simian adenovirus 7. J Gen Virol. 2011;92:1749–1753. doi: 10.1099/vir.0.032300-0. [DOI] [PubMed] [Google Scholar]
- Roy S, Sandhu A, Medina A, Clawson DS, Wilson JM. Adenoviruses in fecal samples from asymptomatic rhesus macaques, United States. Emerg Infect Dis. 2012;18:1081–1088. doi: 10.3201/eid1807.111665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Vandenberghe LH, Kryazhimskiy S, Grant R, Calcedo R, Yuan X, Keough M, Sandhu A, Wang Q, Medina-Jaszek CA, Plotkin JB, Wilson JM. Isolation and characterization of adenoviruses persistently shed from the gastrointestinal tract of non-human primates. PLoS Pathog. 2009;5:e1000503. doi: 10.1371/journal.ppat.1000503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saad RS, Demetris AJ, Lee RG, Kusne S, Randhawa PS. Adenovirus hepatitis in the adult allograft liver. Transplantation. 1997;64:1483–1485. doi: 10.1097/00007890-199711270-00021. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Treacy A, Carr MJ, Dunford L, Palacios G, Cannon GA, O'Grady A, Moran J, Hassan J, Loy A, Connell J, Devaney D, Kelehan P, Hall WW. First report of sudden death due to myocarditis caused by adenovirus serotype 3. J Clin Microbiol. 2010;48:642–645. doi: 10.1128/JCM.00815-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh MP, Chintakuntlawar A, Robinson CM, Madisch I, Harrach B, Hudson NR, Schnurr D, Heim A, Chodosh J, Seto D, Jones MS. Evidence of molecular evolution driven by recombination events influencing tropism in a novel human adenovirus that causes epidemic keratoconjunctivitis. PLoS One. 2009;4:e5635. doi: 10.1371/journal.pone.0005635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wevers D, Metzger S, Babweteera F, Bieberbach M, Boesch C, Cameron K, Couacy-Hymann E, Cranfield M, Gray M, Harris LA, Head J, Jeffery K, Knauf S, Lankester F, Leendertz SA, Lonsdorf E, Mugisha L, Nitsche A, Reed P, Robbins M, Travis DA, Zommers Z, Leendertz FH, Ehlers B. Novel adenoviruses in wild primates: a high level of genetic diversity and evidence of zoonotic transmissions. J Virol. 2011;85:10774–10784. doi: 10.1128/JVI.00810-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wold WSM, Horwitz MS. Adenoviruses. In: Knipe DM, Howley PM, editors. Fields Virology. 5th ed. Philadelphia: Lippincott–Williams & Wilkins; 2007. pp. 2395–2436. [Google Scholar]
- Wolfe ND, Dunavan CP, Diamond J. Origins of major human infectious diseases. Nature. 2007;447:279–283. doi: 10.1038/nature05775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood DJ. Adenovirus gastroenteritis. Br Med J (Clin Res Ed) 1988;296:229–230. doi: 10.1136/bmj.296.6617.229-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano S, Tokino T, Yasuda M, Kaneuchi M, Takahashi M, Niitsu Y, Fujinaga K, Yamashita T. Induction of transformation and p53-dependent apoptosis by adenovirus type 5 E4orf6/7 cDNA. J Virol. 1999;73:10095–10103. doi: 10.1128/jvi.73.12.10095-10103.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]