Summary
LncRNAs are known to regulate a number of different development and tumorigenic processes. Here we report a role for lncRNA BCAR4 in breast cancer metastasis that is mediated by chemokine-induced binding of BCAR4 to two transcription factors with extended regulatory consequences. BCAR4 binding of SNIP1 and PNUTS in response to CCL21 releases the SNIP1's inhibition of p300-dependent histone acetylation that in turn enables the BCAR4-recruited PNUTS to bind H3K18ac and relieve inhibition of RNA Pol II via activation of the PP1 phosphatase. This mechanism activates a noncanonical Hedgehog/GLI2 transcriptional program that promotes cell migration. BCAR4 expression correlates with advanced breast cancers and therapeutic delivery of Locked Nucleic Acids (LNAs) targeting BCAR4 strongly suppresses breast cancer metastasis in mouse models. The findings reveal a disease-relevant lncRNA mechanism consisting of both direct coordinated protein recruitment and indirect regulation of transcription factors.
Keywords: Breast Cancer Metastasis, Hedgehog Signaling, long noncoding RNA, BCAR4, GLI2 Phosphorylation, Transcription, H3K18 acetylation, Locked Nucleic Acid
Introduction
Emerging evidence has purported long noncoding RNA (lncRNA) as a new class of players involved in the development and progression of cancer (Fatica and Bozzoni, 2014). However, the regulatory roles played by lncRNAs in breast cancer-associated aberrant signaling pathways/transcriptional programs are not completely understood. LncRNAs exert their regulatory functions through specific interactions with proteins including epigenetic modifiers, transcriptional factors/co-activators and RNP complexes (Rinn and Chang, 2012). The specific lncRNA-protein interactions could be mediated by canonical RNA-binding domains (RBDs) (Lunde et al., 2007) or non-canonical RBDs including tryptophan-aspartic acid 40 (WD40) domain and RNA-binding domain abundant in Apicomplexans (RAP) demonstrated by recent mRNA interactome capture methodology (Castello et al., 2012). Therefore, it is of great interest to uncover new functions of lncRNAs by dissecting lncRNA-protein interactions mediated by noncanonical RBDs in certain biological processes.
The aberrant activation of the hedgehog signaling pathway in breast cancer has been connected with increased expression of the transcription factor, glioma-associated oncogene homolog 1/2 (GLI1/2) (ten Haaf et al., 2009). GLI1/2-dependent target gene transcription has been shown to be involved in tumor cell growth and metastasis in solid tumors (Rubin and de Sauvage, 2006). However, GLI-target transcription might be activated in the absence of the hedgehog ligand Sonic Hedgehog (SHH), especially in triple-negative breast cancer (TNBC) (Hui et al., 2013), suggesting that other mechanisms/regulators may regulate the activity of the GLI transcription factor. The direct binding of lncRNAs to transcription factors (Geisler and Coller, 2013) led us to speculate that the association of transcription factor GLI with lncRNAs may function in regulating GLI-dependent transcriptional program essential for breast cancer progression and metastasis.
The lncRNAs implicated in breast cancer represent a promising class of therapeutic targets. Targeting noncoding RNAs by using Locked Nucleic Acids (LNA)-based antisense oligonucleotides strategy has been a longstanding interest (Dias and Stein, 2002), with several successful applications in targeting miRNAs in cancer (Ling et al., 2013). However, therapeutic targeting of lncRNA has not been well documented for breast cancer. Thus, we aimed to determine the therapeutic potential of targeting breast cancer-upregulated lncRNAs by a LNA-based antisense oligonucleotides strategy.
Here, we report the identification of a signaling pathway that is triggered by CCL21 and mediated by citron (rho-interacting, serine/threonine kinase 21) (CIT) kinase to phosphorylate the transcriptional factor GLI2, which regulates target gene expression in breast cancer cells. The lncRNA BCAR4 is required for phospho-GLI2 dependent gene activation via its direct interaction with Smad nuclear-interacting protein 1 (SNIP1) and Serine/threonine-protein phosphatase 1 regulatory subunit 10 (PPP1R10, also known as PNUTS). Mechanistically, the BCAR4-SNIP1 binding releases the inhibitory role of SNIP1 on p300 histone acetyltransferase (HAT) activity, leading to the acetylation of histones including a novel mark, H3K18ac, on the promoters of GLI2 target transcription units. The acetylated H3K18 can be further recognized by PNUTS, which is recruited to the promoters of GLI2 target genes by BCAR4, to attenuate the protein's inhibitory effect on the enzymatic activity of PP1, leading to hypophosphorylation of RNA polymerase II at Ser5. Elevated BCAR4 expression correlated with higher metastatic potential and shorter survival time of breast cancer patients, whereas it's therapeutic inhibition by LNA displays in vivo efficacy against metastasis. Our findings have provided supporting evidence for the regulatory roles played by lncRNAs in the progression of aggressive breast cancers. Broadly, our results of the therapeutic effectiveness of BCAR4 LNA against breast cancer metastasis document an example to show the pharmacologic value of lncRNA in human cancer and other diseases.
Results
BCAR4 Correlates with Advanced Breast Cancer and Regulates GLI-mediated Transcription
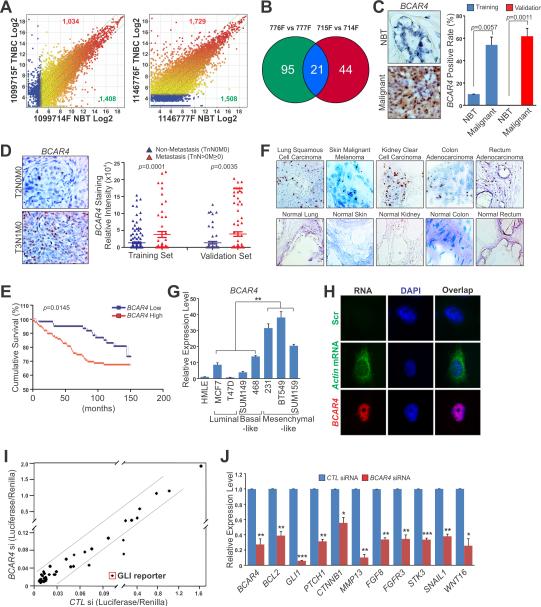
To identify breast cancer-relevant lncRNAs, we profiled the expression of lncRNAs in two stage III breast cancer tissues and their paired adjacent noncancerous tissues (Figure S1A) by LncRNA Array 3.0 (ArrayStar). An average of 1,381 up-regulated lncRNAs (range from 1,034 to 1,729) and 1,458 down-regulated lncRNAs (range 1,408–1,508) with significantly differential expression (≥3.0-fold) were identified (Figure 1A; Table S1). We further compared the lncRNA expression levels between breast cancer tissues and their paired adjacent normal tissues based on the NCBI RefSeq database (which contains 3,991 human lncRNAs with annotated NR accession number), identifying 65 and 116 up-regulated lncRNAs in two patient cases, respectively (≥4.0-fold) (Figure 1B). Among these lncRNAs, 21 were consistently up-regulated in both patient samples, of which BCAR4, initially identified through genetic screening as a novel gene involved in tamoxifen resistance in breast cancers (Meijer et al., 2006), showed the most up-regulation (LogFC: 15.9 and 16.1, respectively) (Figures S1B and S1C).
Figure 1. BCAR4 Correlates with Breast Cancer Metastasis.
(A) Scatter plots of lncRNAs significantly up-regulated (red) or down-regulated (green) in two pairs of TNBC tissues compared to the matched adjacent normal tissues (NBT). The X- and Y-axes: averaged normalized signal values (Log2 scaled); green lines: fold changes=4.
(B) Commonly up-regulated lncRNAs in two pairs of TNBC compared to NBT.
(C) RNAScope® detection of BCAR4 expression in human breast cancer and adjacent normal tissues. Left panel: representative images; Right panel: statistical analysis of training set (10 normal tissues vs. 222 cancer tissues) and validation set (10 normal tissues vs. 160 cancer tissues).
(D) RNAScope® detection of BCAR4 expression in Non-metastasis (TnN0M0) vs. Metastasis (TnN>0M≥0) breast cancer tissue. Left panel: representative images; Right panel: statistical analysis of training set (167 Non-metastasis vs. 55 Metastasis) and validation set (66 Non-metastasis vs. 94 Metastasis).
(E) Kaplan-Meier survival analysis of BCAR4 expression in breast cancer patients (n=160).
(F) RNAscope® detection of BCAR4 expression in multiple human tissues.
(G) RT-qPCR detection of BCAR4 expression in a panel of cell lines.
(H) Nuclear localization of BCAR4 detected by RNA FISH in MDA-MB-231 cells.
(I) Identification of signal pathways affected by BCAR4 knockdown in MDA-MB-231 cells. The X- and Y-axes: normalized ratio of firefly/Renilla luciferase activities.
(J) RT-qPCR detection of GLI-target genes expression. Error bars, S.E.M. of three independent experiments (*p<0.05, **p<0.01 and ***p<0.001).
We first performed RNA in situ hybridization on breast cancer tissue microarrays (clinicopathological features listed in Table S2) using RNAScope® 2.0 HD technology to examine the potential correlation of BCAR4 with breast cancer. In a training set of breast cancer tissue microarrays containing 232 cases, BCAR4 exhibited positive staining only in 10% of the normal breast tissues, while 54.10% of breast cancer tissues showed positive BCAR4 expression (p=0.0057) (Figure 1C). In a validation set containing 170 cases, none of 10 normal adjacent breast tissues showed detectable BCAR4 expression but 61.88% of breast cancer tissues exhibited positive BCAR4 staining (p=0.0011) (Figure 1C). Furthermore, breast cancer at advanced lymph-node metastasis stage (TnN>0M≥0) showed increased BCAR4 expression compared to those early stage tumor with no lymph-node metastasis (TnN0M0) (p=0.0001, training set; p=0.0035, validation set) (Figure 1D). Elevated BCAR4 expression also significantly correlates with shorter survival time of breast cancer patients (n=160, p=0.0145) (Figure 1E). We further analyzed breast cancer database in Oncomine, finding that BCAR4 expression not only correlates with breast cancer but also with triple negativity, lymph-node metastasis and 5 years recurrence (Figure S1D). Oncomine database also showed significant correlation of BCAR4 expression with metastatic prostate cancer, lung cancer, colorectal and rectal cancer (Figure S1D). To confirm this, we employed RNAScope® assay to analyze BCAR4 expression in normal and cancer tissues from multiple organ, observing increased BCAR4 expression in many types of human cancer tissues including colorectal, melanoma and lung cancer, compared to normal tissues (Figure 1F; Table S3). Taken together, these results demonstrated the strong correlation of BCAR4 expression with breast cancer progression and the relevance of elevated BCAR4 expression to human cancer development and progression.
We then examined the expression of BCAR4 in a panel of breast cancer cell lines, finding higher expression of BCAR4 in mesenchymal-like cell lines with metastasis potential compared to epithelial-like cell lines, which are considered as non-metastatic (Figure 1G). We next examined the subcellular localization of BCAR4 by RNA FISH and real-time RT-qPCR analyses on fractionated RNA, finding that the BCAR4 transcript is predominately localized in the nucleus (Figures 1H and S1E). BCAR4 has two major splice variants, full-length transcript (~ 1.3 kb) and an isoform lacking two alternate exons (~680 bp) and our Northern Blot analysis revealed that the full-length isoform was predominately expressed in MDA-MB-231 cells, but truncated isoform barely expressed (Figure S1F). Because the previous report suggested that BCAR4 may encode a small peptide in bovine oocytes (Thelie et al., 2007), we generated an antibody using the predicted translated peptide sequence. However, neither immunoblotting of MDA-MB-231 lysate nor in vitro translation assays showed protein coding potential of BCAR4 (Figure S1G and data not shown).
We next analyzed the effect of BCAR4 knockdown on activation of key signaling pathways in breast cancer cells using Cignal Finder™ 45-Pathway Reporter Array, finding that either siRNA or LNA efficiently depleted BCAR4 expression (Figures S1H and S1I) and knockdown of BCAR4 dramatically inhibited GLI reporter luciferase activity but no other transcription factor reporters (Figure 1I). qRT-PCR analysis confirmed decreased expression of endogenous GLI target genes with BCAR4 knockdown (Figure 1J). These data suggest the potential role of BCAR4 in mediating the GLI-dependent hedgehog signaling pathway in breast cancer cells.
Identification and Biochemical Characterization of BCAR4-associated Proteins
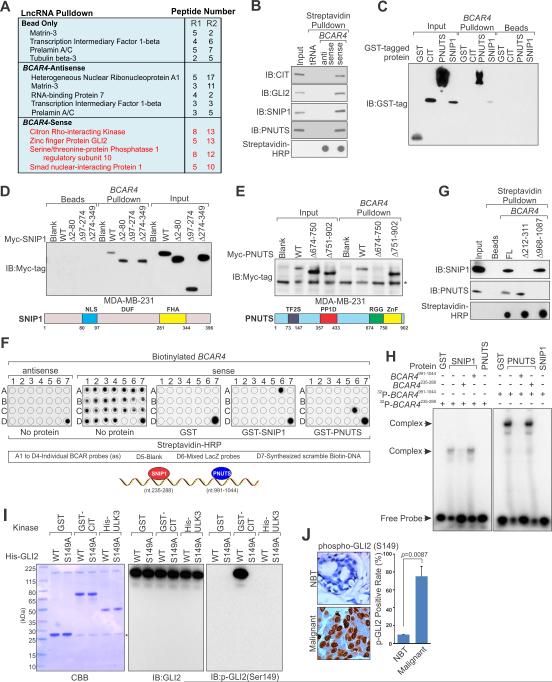
Through RNA pulldown followed by Mass-spectrometry (MS) analysis, we identified that in vitro-transcribed biotinylated BCAR4 sense transcript associated specifically with CIT kinase, GLI2, SNIP1 and PNUTS, even under high stringency wash conditions. However, the antisense transcript of BCAR4 associated with some general RNA-binding proteins that were also bound by the beads (Figures S2A and 2A; Table S4). Of note, in one of two biological repeats of RNA-pulldown experiment, we observed the relative abundant association of BCAR4 with heterogeneous nuclear ribonuclearprotein, which have been reported to bind other lncRNAs (Carpenter et al., 2013; Huarte et al., 2010). Furthermore, the MS data indicated the potential phosphorylation of GLI2 at Serine149 (Figure S2B).
Figure 2. Identification and Biochemical Characterization of BCAR4-associated Proteins.
(A) A list of top BCAR4-associated proteins identified by RNA pulldown and MS analysis in MDA-MB-231 cells: R1 and R2 (biological repeat 1 and 2).
(B and C) Immunoblot (IB) detection of proteins retrieved by in vitro transcribed biotinylated BCAR4 from MDA-MB-231 cell lysates (B) and indicated recombinant proteins (C).
(D and E) IB detection of Myc-tagged SNIP1 (D) and PNUTS (E) (wt vs. domain truncation mutants) retrieved by in vitro transcribed biotinylated BCAR4. Lower panels: graphic illustration of the domain structure of SNIP1 (D) or PNUTS (E).
(F) In vitro RNA-protein binding followed by dot-blot assays. Bottom panel: schematic illustration of the BCAR4 sequence motifs that is recognized by SNIP1 and PNUTS, respectively.
(G) IB detection of proteins retrieved by in vitro transcribed biotinylated BCAR4 (wt vs. Δ212-311 and Δ968-1087) from MDA-MB-231 cell lysates.
(H) EMSA assay of recombinant SNIP1 and PNUTS binding to BCAR4 nt. 235-288 and nt. 991-1044 respectively.
(I) In vitro kinase assay showing CIT-mediated phosphorylation of GLI2 (wt vs. S149A). *: unspecific band.
(J) IHC staining of phospho-GLI2 (S149) in human breast cancer and adjacent normal tissues. Left panel: representative image. Right panel: statistics analysis based on (10 normal tissues vs. 222 cancer tissues).
The RNA pulldown assays with cell lysate further confirmed the specific association of BCAR4 with the proteins identified by MS analysis (Figure 2B). In vitro RNA-protein binding assay revealed that only PNUTS and SNIP1 directly interact with BCAR4 (Figures 2C and S2C). Protein domain mapping studies demonstrated that BCAR4 binds the 97-274 a.a. region of SNIP1 and 674-750 a.a. region of PNUTS, respectively (Figures 2D and 2E). The 97-274a.a. region of SNIP1 encodes a domain known as the Domain of Unknown Function (DUF) and has been suggested to bind miRNA (Yu et al., 2008), which is consistent with our observation that the DUF of SNIP1 serves as the RNA binding domain for BCAR4. PNUTS also has an RNA binding motif, the 674-750a.a. region known as RGG-box (Kim et al., 2003). To further understand the BCAR4-protein interactions in vivo, we performed immunoprecipitation using antibodies against CIT, GLI2, SNIP1 and PNUTS respectively under the condition of BCAR4 knockdown (Figures S2D and S2E), finding that knockdown of BCAR4 impaired the interaction of PNUTS with proteins CIT, GLI2 and SNIP1, but had minimal effect on the association of CIT, GLI2 and SNIP1 with each other (Figure S2E). Given the observation that only SNIP1 and PNUTS directly bound to BCAR4 (see Figure 2C), our data suggest that SNIP1 mediates the association of CIT and GLI2 with BCAR4 and that SNIP1 and PNUTS bind distinct regions of BCAR4.
To map the BCAR4 sequence motifs responsible for SNIP1 and PNUTS binding, we performed an in vitro RNA pulldown followed by dot-blot assay (Yang et al., 2013). The motif sequence of BCAR4 bound/protected by SNIP1 and PNUTS was identified to encompass 235TGT...GGA288 and 991GTT...ATA1044, respectively (Figure 2F). However, the GST protein showed no specific binding to any region of BCAR4 (Figure 2F). Deletion of the corresponding sequence of BCAR4 (Δ212-311) abolished its interaction between SNIP1 with no effect on PNUTS binding (Figure 2G). Deletion of the motif sequence Δ968-1087 of BCAR4 abolished its interaction with PNUTS, but not SNIP1 (Figure 2G). Electrophoretic mobility-shift assays (EMSA) were further used to confirm the direct binding of BCAR4 with SNIP1 and PNUTS. Incubation of the BCAR4 RNA probe (nt 235-288) and (nt 991-1044) with recombinant SNIP1 and PNUTS, respectively, resulted in specific gel retardation (Figure 2H). Under these conditions, no shift was observed when the corresponding cold probes were used (Figure 2H). We, therefore, conclude that BCAR4 directly bind to SNIP1 and PNUTS via two distinct regions.
Given MS data showing that GLI2 is phosphorylated at Ser149 and associates with CIT kinase (see Figures 2A and S2B), we reasoned that CIT may serve as a kinase to phosphorylate GLI2. In vitro kinase assay indicated that bacterially-expressed wild type GLI2 was phosphorylated by CIT, but not S149A mutant (Figure S2F). ULK3 served as the positive control due to its reported ability to phosphorylate GLI (Maloverjan et al., 2010). In vitro RNA-protein binding assay using biotinylated BCAR4 and GLI2 proteins phosphorylated by CIT in vitro showed no interaction (Figure S2G).
To investigate the role of GLI2 Ser149 phosphorylation in vivo, we generated rabbit polyclonal antibodies that specifically recognized Ser149-phosphorylated GLI2 referred to as p-GLI2 (Ser149) antibody, which specifically detected bacterially-purified GLI2 protein that phosphorylated by CIT in vitro, with minimal reactivity towards GLI2 phosphorylated by ULK3 (Figure 2I). We conclude that p-GLI2 (Ser149) antibody specifically recognizes CIT-mediated Ser149 phosphorylation of GLI2. Next, we evaluate the level of phospho-GLI2 in breast cancer by immunohistochemistry (IHC) analysis of clinical tumor specimens, finding higher p-GLI2 (Ser149) levels in invasive breast cancer tissues compared with adjacent normal tissues (p=0.0087) (Figure 2J). Our IHC staining further revealed increased p-GLI2 (Ser149) level in multiple cancer types compared to their corresponding normal tissues (Figure S2H; Table S5). IHC analysis also revealed higher CIT expression in invasive breast cancer compared with adjacent normal breast tissues (p=0.0055) (Figure S2I) and the staining of phosphorylated GLI2 strongly correlated with that of BCAR4 and CIT staining (Data not shown). Taken together, we identified and characterized that BCAR4 binds a protein complex containing SNIP1, PNUTS, phosphorylated GLI2 and CIT via its direct interaction with SNIP1 and PNUTS.
CCL21 Induces GLI2 Ser149 Phosphorylation and Nuclear Translocation of Phosphorylated GLI2
The CIT kinase-mediated GLI2 phosphorylation prompted us to investigate whether this phosphorylation could be triggered in MDA-MB-231 cells by hedgehog signaling. Surprisingly, although the ligand SHH activated hedgehog signaling in Daoy cells evidenced by stimulated SHH gene induction as previously reported (Wang et al., 2012), minimal effect was observed in MDA-MB-231 cells (Figure S3A) and no phosphorylated GLI2 was detected (data not shown), suggesting that a noncanonical hedgehog signaling pathway, involving Ser149-phosphorylated GLI2, may exist in breast cancer.
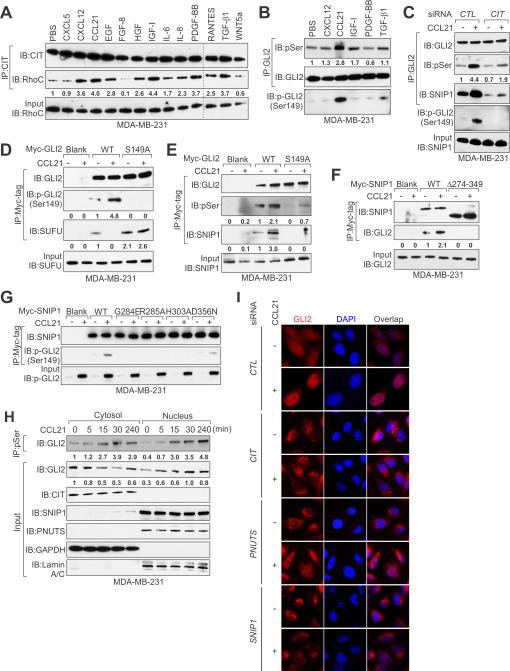
We then explored whether extracellular signals that activate CIT kinase could also trigger GLI2 phosphorylation in breast cancer cells. Given that CIT kinase can be activated by GTPase Rho proteins (Madaule et al., 1998), we first screened the CIT-Rho interaction in breast cancer cells. Although CIT kinase is constitutively associated with RhoA as previously reported (Gai et al., 2011), the presence of Rho activator specifically triggered the interaction between RhoC and CIT kinase (Figure S3B). Then, we hypothesized that the RhoC-activating stimulus may activate CIT kinase. Indeed, we screened 13 known growth factors/cytokines/chemokine involved in RhoC activation and breast cancer metastasis (Favoni and de Cupis, 2000; Kakinuma and Hwang, 2006), finding that CXCL12, CCL21, IGF-I, PDGF-BB, and TGF-β1 enhanced the interaction between RhoC and CIT (Figure 3A). The same stimuli induced activation of CIT kinase indicated by phosphorylation of MLC, a classic CIT kinase substrate (Yamashiro et al., 2003), with CCL21 exhibiting the highest induction (Figure S3C). We then tested the phosphorylation of GLI2 in MDA-MB-231 cells treated with CXCL12, CCL21, IGF-1, PDGF-BB, and TGF-β1, finding that CCL21 dramatically induced Ser149 phosphorylation of GLI2 (Figure 3B), which was significantly reduced by CIT knockdown (Figure 3C). Consistently with previous finding that CCL21-CCR7 autocrine signaling is critical for breast cancer metastasis (Muller et al., 2001), treatment of MDA-MB-231 cells with either neutralizing anti-CCL21 or anti-CCR7 antibodies inhibited basal or CCL21-induced GLI2 phosphorylation (Figures S3D and S3E). CCL21 treatment also dramatically induced GLI2 Ser149 phosphorylation in a panel of additional cancer cell lines, ruling out the possibility of cell line-specific effect (Figure S3F).
Figure 3. Identification of A Noncanonical Hedgehog Signaling Pathway Mediated by CCL21/CIT/phospho-GLI2 Signaling Axis.
(A and B) Immunoprecipitation (IP) and IB detection of CIT-RhoC interactions (A) and GLI2 phosphorylations (B) in cells treated with indicated growth factors, cytokines or chemokines.
(C) IP and IB of GLI2 phosphorylations in cells transfected with indicated siRNAs followed by CCL21 treatment.
(D and E) IP and IB detection of GLI2-SUFU (D) or GLI2-SNIP1 (E) interactions in MDA-MB-231 cells transfected with GLI2 (wt vs. S149A) followed by CCL21 treatment.
(F and G) IP and IB detection of GLI2-SNIP1 interactions in cells transfected with SNIP1 (wt vs. Δ274-349) (F) or (wt vs. FHA domain point mutants) (G) followed by CCL21 treatment.
(H and I)) IP and IB (H) and Immunofluorescence (I) detection of phospho-GLI2 nuclear translocation in cells treated with CCL21 treatment at different time points (H) or transfected with indicated siRNAs followed by CCL21 treatment (I).
See also Figure S3.
Next, we investigated the functional consequence of Ser149 phosphorylation on GLI2. In the cytoplasm, GLI is associated with the Suppressor of Fused Homolog (SUFU), which regulates the cellular localization of GLI (Dunaeva et al., 2003). We performed coimmunoprecipitation experiments and observed that CCL21 treatment induced dissociation between GLI2 and SUFU (Figure S3G), while the exogenously expressed GLI2 S149A mutant failed to release from SUFU in response to CCL21 (Figure 3D). Given that SNIP1, which is in the same complex with GLI2 (see Figure 2A), harbors an FHA domain that recognizes phosphoserine/threonine, we hypothesized that Ser149 phosphorylation of GLI2 is required for its interaction with SNIP1 via the FHA domain. Indeed, either knockdown of CIT or introduction of S149A mutant reduced CCL21-induced interaction between GLI2 and SNIP1 (Figures 3C and 3E). Consistently, deletion or point mutation of amino acids that are critical for FHA domain function (Durocher et al., 2000) also abolished SNIP1's interaction with phosphorylated GLI2 (Figures 3F and 3G). We then performed nuclear fractionation experiments, finding that phosphorylated GLI2 translocated to the nucleus upon CCL21 treatment; whereas CIT, SNIP1 and PNUTS did not exhibit relocation (Figure 3H). The phospho-GLI2 specific antibody also exhibited nuclear staining patterns in breast cancer tissue samples (see Figure 2J). Knockdown of CIT or SNIP1 abolished CCL21-induced nuclear translocation of GLI2 (Figure 3I). In accordance with this, GLI2 S149A mutant failed to translocate into the nucleus upon CCL21 treatment (Figure S3H). Our findings reveal a CCL21/CIT kinase/phospho-GLI2/SNIP1 signaling cascade in breast cancer cells, which may represent a noncanonical hedgehog pathway.
BCAR4 is Required for Transcription Activation of Phospho-GLI2-dependent Target Genes in Breast Cancer Cells
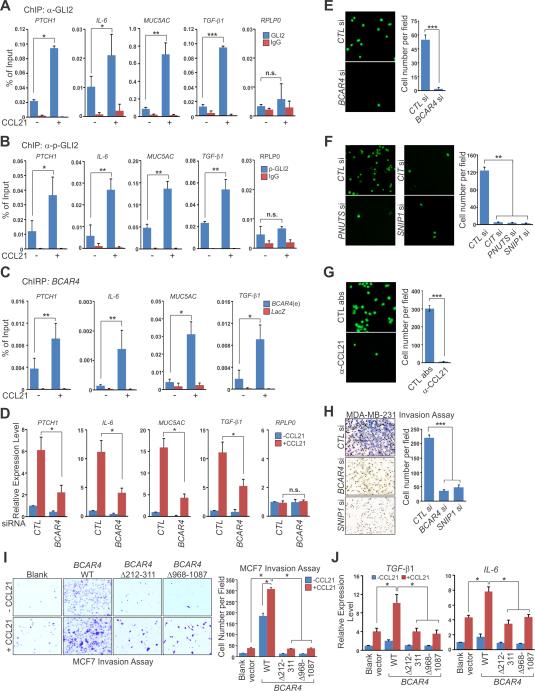
To test if CCL21/CIT/SNIP1 signaling axis-mediated phospho-GLI2 nuclear translocation leads to the activation of GLI target genes, we performed a ChIP assay using antibodies against GLI2 or phospho-GLI2, finding that Ser149 phosphorylated GLI2 was present on the promoters of several well-established GLI target genes PTCH1, IL-6, MUC5AC and TGF-β1, but not on the promoter of a non-GLI target gene, RPLP0 (Figures 4A and 4B). We then performed a ChIRP assay to examine the genomic occupancy of BCAR4, finding that in response to CCL21 treatment, BCAR4 was recruited to the promoters of PTCH1, IL-6, MUC5AC, and TGF-β1 (Figures 4C, S3I and S3J). Consistently, either knockdown of BCAR4 or overexpression of GLI2 S149A mutant dramatically impaired CCL21-induced expression of PTCH1, IL-6, MUC5AC, and TGF-β1 genes (Figure 4D and data not shown).
Figure 4. BCAR4 Is Required for CCL21-triggered, phospho-GLI2-mediated Gene Activation and Cell Migration.
(A-C) ChIP-qPCR detection of GLI2 (A), phospho-GLI2 (B) or ChIRP-qPCR detection of BCAR4 (C) occupancy on the promoters of selected GLI2 target genes in MDA-MB-231 cells treated with CCL21. RPLP0 served as a non-GLI2 target gene control (A and B).
(D) RT-qPCR detection of GLI2 target genes expression in MDA-MB-231 cells transfected with control or BCAR4 siRNA followed by CCL21 treatment.
(E to G) Cell migration assays in MDA-MB-231 cells transfected with indicated siRNA (E and F) or treated with CCL21 neutralization antibody (G).
(H) Matrigel cell invasion assay in cells transfected with indicated siRNAs. Left panel: representative images; right panel: statistical analysis.
(I and J) Matrigel cell invasion assay (I) or RT-qPCR detection of GLI2 target genes (J) in MCF7 cells electroporated with indicated BCAR4 expression vectors followed by CCL21 treatment.
Error bars, SEM of three independent experiments (*p<0.05, **p<0.01 and ***p<0.001).
See also Figure S4.
One of the major biological roles of GLI is to modulate the gene expression related to cell migration and invasion (Feldmann et al., 2007). Thus, we examined the effect of GLI2, BCAR4, and other BCAR4 bound proteins on breast cancer cell invasion and migration. The treatment of MDA-MB-231 cells with validated siRNAs against BCAR4, CIT, SNIP1, or PNUTS or neutralizing antibody against CCL21 all dramatically inhibited cell migration (Figures 4E-4G) and invasion (Figures 4H and data not shown) but did not affect cell proliferation (Figure S4A). Consistently, stable knockdown of BCAR4 by shRNAs in MDA-MB-231 LM2 cells reduced migration and invasion properties of these cells (Figures S4B-S4D). We also tested if BCAR4 is critical for migration and invasion of those metastatic cancer cell lines that respond to CCL21 treatment (see Figure S3F). Our data showed that while knockdown of BCAR4 had no effect on proliferation of HCT116, H1299, HepG2 and Hey8 cells (Figures S4E and S4F), the migration and invasion of these cells were significantly reduced (Figures S4G, S4H and data not shown). In addition, CCL21-induced GLI2 target genes expression in these cell lines was inhibited by BCAR4 knockdown (Figures S4I, S4J and data not shown).
Given that BCAR4 is critical for metastasis potential of cancer cells and our observation of lower BCAR4 expression level in non-metastatic breast cancer cell lines compared to metastatic breast cell lines (see Figure 1G), we reasoned that overexpression of BCAR4 in a nonmetastatic cell line may increase its metastasis potential. MCF-7 is a non-metastatic breast cancer cell line but expresses the CCR7, the receptor for CCL21 (Muller et al., 2001). Indeed, stimulation of MCF-7 cells with CCL21 modestly enhanced their invasion (Figure 4I). However, overexpression of full-length BCAR4 but not the deletion mutants abolishing SNIP1 or PNUTS binding in MCF-7 cells (Figure S4K) increased the invasion and GLI2 target genes expression even under the basal condition (Figures 4I, 4J and S4L), which was not due to cell proliferation effect (Figure S4M). These data strongly argue the important role of BCAR4 in the phospho-GLI2-mediated transcription activation of a subset of genes, which may contribute to breast cancer cell migration and invasion.
BCAR4 Binds SNIP1 and Release the Inhibitory Effect of SNIP1 on p300 HAT Activity
We next investigated the molecular mechanism by which BCAR4 regulates GLI2 target genes expression. Considering that BCAR4 directly interacts with SNIP1 in vitro, we explored whether this interaction is functionally important in vivo by examining the SNIP1-BCAR4 interaction by RNA Immunoprecipitation (RIP) assay, finding that in response to CCL21 treatment, SNIP1 bound to BCAR4 in multiple cancer cell lines (Figures S5A-S5C). As a control, no interaction between SNIP1 and NEAT2, an abundant nuclear lncRNA, was observed (Figures S5A-S5C). As expected, deletion of the 97-274 a.a. region abolished SNIP1-BCAR4 interaction (Figure 5A), which is consistent with our previous observation that the DUF domain of SNIP1 is required for SNIP1-BCAR4 interaction (see Figure 2D). Surprisingly, deletion of the FHA domain (region 274-349 a.a.) of SNIP1 led to constitutive SNIP1-BCAR4 interaction (Figures 5A and S5D), suggesting that binding to phospho-serine/threonine via its FHA domain, is required for SNIP1's subsequent interaction with BCAR4, possibly through a mechanism involving the conformational change of SNIP1 upon phospho-GLI2 binding. Indeed, FHA domain mutants of SNIP1 all failed to interact with BCAR4, while wild type SNIP1 along with the D356N mutant, which exhibits no effect on phospho-GLI2 binding, was able to bind BCAR4 (Figure 5B). These data suggest that SNIP1's FHA domain may block the DUF domain, preventing SNIP1-BCAR4 interaction. Upon stimulation, the FHA domain recognizes phospho-Ser149 of GLI2, which causes conformational changes that may expose the DUF domain for BCAR4 binding.
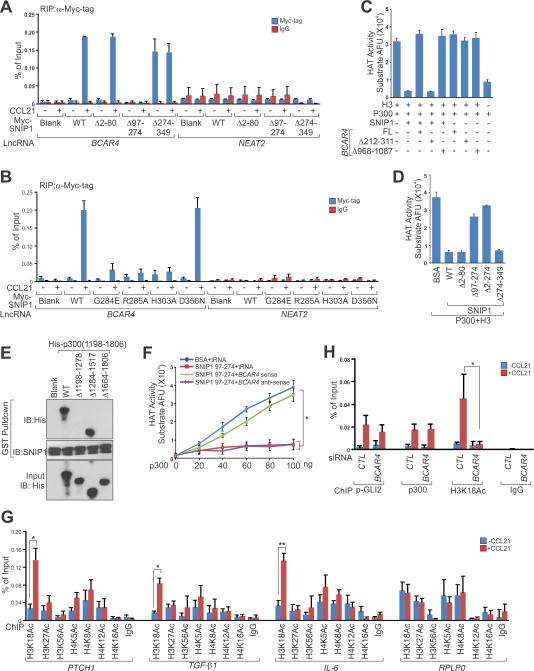
Figure 5. Signal-induced BCAR4-SNIP1 Interaction Attenuates the Inhibitory Effect of SNIP1 on p300 HAT Activity.
(A and B) RIP-qPCR detection of the indicated RNAs retrieved by Myc-specific antibody in MDA-MB-231 cells electroporated with indicated vectors followed by CCL21 treatment.
(C) In vitro HAT activity assays of p300 in the presence of wt SNIP1, full-length (FL) BCAR4 and their corresponding mutants as indicated.
(D) In vitro HAT activity assays of p300 in the presence of wt SNIP1 and its corresponding mutants as indicated.
(E) IB detection of the interaction between SNIP1 (a.a. 97-274) and p300 (a.a. 1198-1806) wt or truncations.
(F) In vitro HAT activity assays of p300 in the presence of SNIP1 a.a. 97-274 and BCAR4 sense or antisense RNAs.
(G) ChIP-qPCR detection of acetylated histone marks occupancy on the promoters of selected GLI2 target genes in MDA-MB-231 cells treated with CCL21.
(H) ChIP-qPCR detection of phospho-GLI2, p300 and H3K18Ac occupancy on PTCH1 promoter in MDA-MB-231 cells transfected with indicated siRNAs followed by CCL21 treatment.
Error bars, SEM of three independent experiments (*p<0.05 and **p<0.01).
See also Figure S5.
SNIP1 has been reported to interact with p300 and potentially regulates p300-dependent gene transcription (Kim et al., 2000). Although immunoprecipitation of SNIP1 confirmed its interaction with p300, the interaction was not affected by deprivation of BCAR4 (Figure S5E). Deletion of either DUF domain of SNIP1 (region 97-274a.a.) or the BCAR4 SNIP1 binding motif (nt 212-311) exhibited minimal effect on SNIP1-p300 interaction (Figures S5F and S5G). We then examined the HAT activity of p300 in the presence of SNIP1 and/or BCAR4. Surprisingly, the HAT activity of p300, was strongly inhibited by recombinant SNIP1, but could be rescued by in vitro transcribed BCAR4 RNA (Figure 5C). This rescue was dependent on the interaction between BCAR4 and SNIP1's DUF domain because the presence of BCAR4 alone had no effect on the HAT activity of p300. Moreover, deletion of BCAR4's SNIP1 binding motif (nt 212-311) abolished the rescue of p300's HAT activity (Figure 5C). Therefore, our data indicated that the interaction between SNIP1 and BCAR4 released the inhibitory role of SNIP1 on the HAT activity of p300.
Although it has been suggested that SNIP1 regulates the p300-dependent transcription of multiple signaling pathways (Fujii et al., 2006; Kim et al., 2001; Kim et al., 2000), the mechanism is not clear. We mapped the domains of SNIP1 that may interact with p300 and found that while both the N-terminal (2-80 a.a.) and DUF domain (97-274 a.a.) of SNIP1 were required for p300 binding (Figure S5H), the DUF domain of SNIP1 is the minimum region required to inhibit the enzymatic activity of p300 (Figure 5D). By incubating SNIP1 with p300 catalytic unit (a.a. 1198-1806) and derivative truncation mutants, we found that the DUF domain of SNIP1 interact with PHD (a.a. 1198-1278) and CH3 domains (a.a 1664-1806) of p300 catalytic unit, which may interfere with p300's HAT activity (Figure 5E). According to our in vitro observations, the DUF domain also binds BCAR4, raising a possible role of BCAR4 in regulating p300's HAT activity. Indeed, in the presence of BSA and tRNA, p300 exhibited dose-dependent HAT activity which was abolished in the presence of SNIP1 DUF domain alone (Figure 5F). In contrast, in the presence of sense but not antisense BCAR4, p300 HAT activity was largely rescued (Figure 5F). These data suggest that the DUF domain of SNIP1 binds PHD and CH3 domains of p300 to inhibit the HAT activity, while signal-induced binding of BCAR4 to SNIP1 DUF domain releases its interaction with the catalytic domain of p300, leading to the activation of p300.
p300-mediated histone acetylation is critical for transcription activation (Wang et al., 2008). We then screened histone acetylation on GLI2 target gene promoters, finding that H3K18ac, H3K27ac, H3K56ac, H4K8ac, H4K12ac, and H4K16ac were induced by CCL21 treatment in breast cancer cells, with H3K18ac showing the highest level (Figure 5G). Knockdown of BCAR4 abolished CCL21-induced H3K18 acetylation on GLI2 target gene promoters; however, this was not due to reduced recruitment of phosphorylated-GLI2 or p300 to GLI2 (Figure 5H). These findings suggest that BCAR4 activates p300 by binding SNIP1's DUF domain to release the inhibitory effect of SNIP1 on p300, which results in the acetylation of histone marks required for gene activation.
Recognition of BCAR4-dependent Histone Acetylation by PNUTS Attenuates Its Inhibitory Effect on PP1 Activity
Based on our data that the 3’ of BCAR4 interacts with PNUTS in vitro, we next examined this interaction in vivo by RIP experiments. We found that PNUTS constitutively interacts with BCAR4 via its RGG domain (Figures S5A-S5C, S6A and 6A), which is consistent with our in vitro data (see Figure 2E). PNUTS functions as a regulatory subunit for PP1, inhibiting the phosphatase activity of PP1 (Kim et al., 2003). As such, we wondered whether BCAR4 could regulate PP1's phosphatase activity via binding PNUTS. The immunoprecipitation assay indicated that knockdown of BCAR4 has minimal effect on PNUTS-PP1A interaction (Figures S1I and S6B). As previously reported (Kim et al., 2003), the phosphatase activity of PP1 was inhibited by PNUTS (Figure S6C). However neither sense nor antisense BCAR4 could rescue PP1's activity (Figure S6D), leading us to explore whether any histone modifications could rescue PP1 activity given that recruitment of the PNUTS/PP1 complex by BCAR4 could possibly activate the transcription of GLI2 target genes.
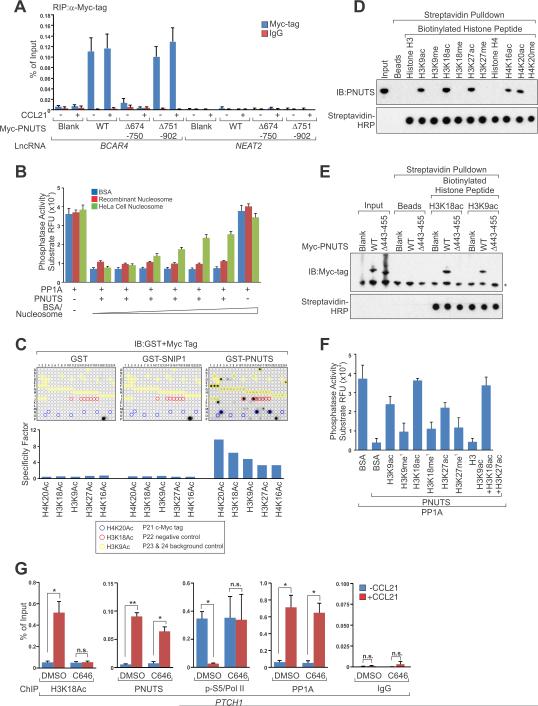
Figure 6. Recognition of BCAR4-dependent Histone Acetylation by PNUTS Attenuates Its Inhibitory Effect on PP1 Activity.
(A) RIP-qPCR detection of the indicated RNAs retrieved by Myc-specific antibody in MDA-MB-231 cells transfected with indicated vectors followed by CCL21 treatment.
(B) In vitro Phosphatase activity assays of PP1A in the presence of indicated proteins or nucleosome.
(C) MODified Histone Peptide Array® detection of histone marks recognition by SNIP1 or PNUTS. Top panel: representative images; bottom panel: binding specificity.
(D and E) IB detection of PNUTS retrieved by biotinylated histone peptides as indicated from lysate of MDA-MB-231 cell (D) or MDA-MB-231 cell electroporated with indicated vectors (E).
(F) In vitro Phosphatase activity assays of PP1A in the presence of PNUTS and modified histones H3 as indicated.
(G) ChIP-qPCR detection of H3K18Ac, PNUTS, Pol II Ser5 (normalized by Pol II occupancy) and PP1A occupancy on PTCH1 promoter in MDA-MB-231 cells pre-treated with C646 followed by CCL21 treatment.
Error bars, SEM of three independent experiments (*p<0.05 and **p<0.01).
See also Figure S6.
Surprisingly, the inhibition of PP1's phosphatase activity by PNUTS was largely rescued by purified nucleosome from HeLa cells but not recombinant nucleosome while neither nucleosome alone affected PP1 activity (Figure 6B), suggesting that modified histones binding is critical to release PNUTS's inhibitory effect on PP1 activity. We then utilized a Modified Histone Peptide Array to test this possibility, finding that PNUTS, but not SNIP1, directly recognized acetylated histones including H4K20ac, H3K18ac, H3K9ac, H3K27ac, and H4K16ac (Figure 6C), which was confirmed by histone peptide pulldown experiments (Figure 6D). A previous study indicated that a minimum region from 445-450 a.a. of PNUTS is required to inhibit the phosphatase activity of PP1 (Kim et al., 2003). We then examined if acetylated histone could also recognize this region, finding that deletion of a.a. 443-455 of PNUTS abolished its interaction with acetylated histone H3 (Figure 6E), suggesting that the inhibitory role of PNUTS, mediated by motif a.a. 443-455, is attenuated in the presence of acetylated histone, leading to activation of PP1 enzymatic activity. Consistently, acetylated, but not methylated, histone peptides specifically rescued PP1 activity from PNUTS inhibition (Figure 6F).
PP1 has been reported to dephosphorylate the Carboxyl-Terminal Domain (CTD domain) of RNA polymerase II at Ser5, which is accumulated at promoter regions of target genes (Komarnitsky et al., 2000; Washington et al., 2002). A recent study showed that depletion of PNUTS in Drosophila results in global hyperphosphorylation of RNA Pol II Ser5, leading to global transcription pause and development defect (Ciurciu et al., 2013). Therefore, we next tested if PNUTS/PP1 regulates phosphorylation of RNA Pol II Ser5, finding that knockdown of PNUTS led to the hyperphosphorylation of RNA Pol II Ser5 (Figures S6E and S6F). We then investigated the functional roles of PNUTS-acetylated histone interaction in regulating the status of RNA Pol II Ser5 phosphorylation in the presence of a p300 inhibitor, C646, which eliminated the histone acetylation as represented by H3K18ac (Figures 6G, S6G and S6H). Our data indicates that CCL21-triggered recruitment of PNUTS and PP1 to the promoters of GLI2 target genes was not affected by p300 inhibitor (Figures 6G, S6G and S6H) and the levels of Pol II Ser5 phosphorylation on these promoters were decreased upon CCL21 treatment (Figures 6G, S6G and S6H). However, the CCL21-induced hypophosphorylation of RNA Pol II Ser5 was abolished in the presence of the p300 inhibitor (Figures 6G, S6G and S6H), suggesting that histone acetylation-dependent PP1 activity modulates RNA Pol II Ser5 phosphorylation level at gene promoter regions. Taken together, the data demonstrate the important roles of BCAR4, through its interaction with SNIP1 and PNUTS, in linking signal-induced acetylation of histone to general transcription machinery during the activation of the GLI2 target genes in breast cancer cells.
BCAR4 as a Potential Therapeutic Target for Breast Cancer Metastasis
To further confirm the functional connection between BCAR4 and breast cancer metastasis, we performed functional rescue experiments in which we depleted BCAR4 by LNA followed by overexpression in MDA-MB-231 cells of either LNA-resistant full-length BCAR4 or truncated mutants defective for SNIP1 or PNUTS binding (see Figures 2F-2H and Figure S7A). In cell motility assays, knockdown of BCAR4 reduced migration and invasion of MDA-MB-231 cells, which could be rescued by re-introduction of full-length, but neither Δ212-311 nor Δ968-1087 truncated form of BCAR4 (Figures S7B and S7C), even though the expression of full-length BCAR4 and truncated forms was equal (Figure S7A), and cell proliferation was not altered (data not shown). Knockdown of BCAR4 also curtailed the expression of GLI2 target genes and re-introduction of full-length BCAR4, but neither Δ212-311 nor Δ968-1087 truncated forms of BCAR4 was able to robustly rescue the induction of these genes (Figures S7D and S7E). Consistently, knockdown of BCAR4 abolished CCL21-induced SNIP1 and PNUTS interaction, while re-introduction of full-length BCAR4, but neither Δ212-311 nor Δ968-1087 truncated forms of BCAR4 was able to robustly rescue the interaction (Figure S7F). These data suggest that BCAR4 exerts a quantitatively-important role in GLI2-dependent target gene activation and cell migration/invasion via its direct interactions with SNIP1 and PNUTS.
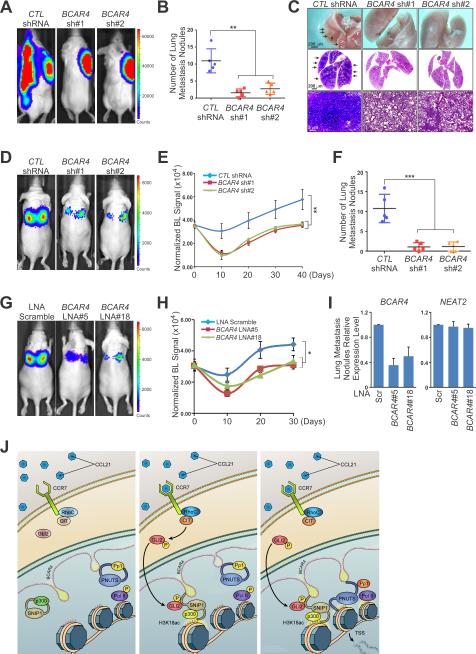
We next set to recapitulate the contribution of BCAR4 to breast cancer metastasis in vivo using highly metastatic MDA-MB-231 LM2 cells harboring shRNA targeting BCAR4, which showed reduced migration and invasion (see Figures S4B-S4D). Bioluminescent imaging (BLI) measurements revealed that mammary gland fat pad injection of MDA-MB-231 LM2 cells harboring control shRNA resulted in lung metastases in NOD/SCID mice while lung metastasis was significantly reduced in two individual groups of mice injected with cells harboring BCAR4 shRNA (Figure 7A), which was confirmed by quantification of lung metastasis nodules (with an average of 11.2 per mouse in control group, and an average of 2 visible metastases per mouse in BCAR4 knockdown groups) and histological examination (Figures 7B and 7C). BCAR4 knockdown had no effect on primary tumor size, tumor cell proliferation or apoptosis (Figures S7G and S7H), indicating that the metastasis suppression phenotype is not secondary to impaired proliferation or apoptosis. However, CD31, a marker for angiogenesis, was significantly downregulated by BCAR4 knockdown (Figure S7H), suggesting that reduced lung metastasis burden is due to defective angiogenesis. Independently, the mice with tail vein injection of BCAR4 knockdown cells rarely developed lung metastases (Figures 7D-7F). Immunohistochemical analyses confirmed efficient inhibition of metastasis (Figure S7I). These data suggest that BCAR4 contribute to breast cancer metastasis and silencing of BCAR4 inhibits lung metastasis in transplantable mouse models.
Figure 7. The Potential Therapeutic Role of BCAR4 in Breast Cancer Metastasis.
(A-C) Representative bioluminescent (BLI) images (A), metastatic nodules numbers in the lungs (B) or isolated lung bright-field imaging (top panel) and H&E staining (middle and bottom panels) (C) of mice with fat pad injection of MDA-MB-231 LM2 cells harboring indicated shRNA. Data are means ± SEM (n=5).
(D-F) Representative BLI images (D), lung colonization (E), and metastatic nodules numbers in the lungs (F) of mice with tail vein injection of MDA-MB-231 LM2 cells harboring indicated shRNA. Data are means ± SEM (n=5).
(G and H) Representative BLI images (G) and lung colonization (H) of mice at day 30 after tail vein injection of MDA-MB-231 LM2 cells followed by intravenously LNA treatment. Data are means ± SEM (n=3).
(I) RT-qPCR detection of BCAR4 expression in sorted GFP positive MDA-MB-231 LM2 cells dissociated from lung metastatic nodules of mice intravenously treated with LNA at day 30 (n=3).
(J) A model for cooperative epigenetic regulation downstream of chemokine signals by lncRNA BCAR4.
To evaluate the potential therapeutic potential of BCAR4, we synthesized LNAs targeting BCAR4. Transfection of LNAs against BCAR4 into MDA-MB-231 cells exhibited strong knockdown efficiency (see Figure S1I) and dramatically affected cell migration and invasion (data not shown). We next examined the therapeutic efficacy of systemically administered in vivo-optimized LNAs in breast cancer metastasis prevention. Of note, two individual LNA treatments significantly reduced lung metastases (Figures 7G and 7H) without notable weight loss (Figure S7J). Importantly, therapeutic LNA-mediated BCAR4 targeting was confirmed by qRT-PCR analysis of lung metastatic nodules (Figure 7I). Taken together, our findings reveal a BCAR4-dependent regulatory network that converges onto a noncanonical hedgehog signaling pathway mediated by phospho-GLI2 to control metastatic initiation and progression in breast cancer.
Discussion
Effective treatment options for breast cancer metastasis, especially for TNBC is not well-established. LncRNA-based mechanisms in breast cancer may represent the crucial nodal points for therapeutic intervention. Our studies have revealed that the lncRNA BCAR4 is highly upregulated in advanced breast cancer patients and contribute to breast cancer metastasis mediated by chemokine-induced binding of BCAR4 to two transcription factors with extended regulatory consequences, licensing the activation of a noncanonical Hedgehog/GLI2 transcriptional program that promotes cell migration (Figure 7J). In a variety of cancer types, including prostate, breast, ovarian, and pancreatic cancers, hedgehog signaling pathways are aberrantly activated, which are critical for tumor progression and invasion. We are tempted to speculate that other lncRNAs in these cancer types recognize covalent modifications of GLI2 or other proteins and exert an analogous function to promote the aberrant cancer signaling pathways, which confers cancer cells the invasiveness and metastatic propensity.
While our data reveal that BCAR4 exerts a quantitatively-important role in chemokine-dependent Hedgehog target gene activation in breast cancer cells, the full mechanisms by which it functions in development remain incompletely defined. BCAR4 is also highly expressed in human oocyte and placenta (Godinho et al., 2011), suggesting its potential roles in development. Interestingly, Hedgehog ligands are expressed in a tissue-specific manner, e.g. Desert Hedgehog (Dhh) expression is specific to sertoli cells of the testes and granulosa cells of ovaries (Varjosalo and Taipale, 2008). These observations indicate that BCAR4 is also critical for GLI-mediated gene expression during development.
The BCAR4 upregulation in breast cancer could be the result of the dysregulation of estrogen receptor (ER). Previous studies have shown that BCAR4 is upregulated in response to tamoxifen treatment of breast cancer cells (Godinho et al., 2011); thus, up-regulation of BCAR4 could be the result of ER down-regulation, as seen in TNBC. It is also possible that BCAR4 expression is regulated at the transcriptional level by certain aberrant oncogenic signaling pathways in breast cancer cells or by gene amplification at the genomic level. Thus, BCAR4 expression may require further investigation.
The targeting of lncRNAs with LNAs in breast cancer has not gained much momentum due to the lack of identification of critical breast cancer-relevant lncRNAs and rigorous investigation of the potential anticancer effects of the modulation of lncRNAs in vivo. The important prognostic capacity of BCAR4 and the robust metastasis suppression by therapeutically delivered LNA targeting BCAR4 documented in our study encourage future development of lncRNA-based cancer therapies for patients at high risk for metastasis -an outcome currently lacking effective chemotherapeutic options.
Experimental Procedures
LncRNA Array v 3.0
Total RNA was extracted from two pairs of fresh frozen infiltrating ductal carcinomas of the breast and their adjacent normal breast tissues. RNA samples were subjected to human genome-wide lncRNA microarray 3.0 analyses at ArrayStar Inc. LncRNA Array data are deposited in the Gene Expression Omnibus database under accession GSE60689. Details are included in Extended Experimental Procedures.
Tissue Specimens
Fresh frozen breast carcinomas and their adjacent normal tissues were purchased from Asterand Inc. Breast cancer tissue microarrays were purchased from Biomax and US BioLab, which were grouped into two sets: training set (BC081120, BR1505a and BR487 from Biomax) and validation set (Bre170Sur-01 from US Biolab). All clinicopathological features of tissue specimens are listed in Table S2.
RNAScope® Assay
The RNAScope® probe targeting BCAR4 was designed and synthesized by Advanced Cell Diagnostics and detection of BCAR4 expression was performed using the RNAscope® 2.0 High Definition (HD)—BROWN Assay according to the manufacturer's instructions (Advanced Cell Diagnostics). The images were acquired with Zeiss Axioskop2 Plus Microscope.
RNA Pulldown and Mass Spectrometry Analysis
Biotin-labeled BCAR4 RNAs were in vitro transcribed with the Biotin RNA Labeling Mix (Roche) and T7 or SP6 RNA polymerase (Ambion) and purified by RNA Clean & Concentrator™-5 (Zymo Research). The cell lysates were freshly prepared using ProteaPrep Zwitterionic Cell Lysis Kit, Mass Spec Grade (Protea®) with Anti-RNase, Protease/ Phosphatase Inhibitor Cocktail, Panobinostat and Methylstat supplemented in the lysis buffer. The BcMag™ Monomer avidin Magnetic Beads (Bioclone) were first prepared according to manufacturer's instructions and then immediately subjected to RNA (20 µg) capture in RNA capture buffer [20 mM Tris-HCl (pH 7.5), 1M NaCl, 1mM EDTA] for 30 minutes at room temperature with agitation. The RNA-captured beads were washed once with NT2 buffer [50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1 mM MgCl2, 0.05% NP-40] and incubated with 30 mg cell lysates diluted in NT2 buffer supplemented with 50 U/mL RNase OUT™, 50 U/mL Superase•IN™, 2 mM dithiothreitol, 30 mM EDTA and Heparin 0.02 mg/ml for 2 hours at 4°C with rotation. The RNA-binding protein complexes were washed sequentially with NT2 buffer (twice), NT2-high salt buffer containing 500 mM NaCl (twice), NT2-high salt buffer containing 1 M NaCl (once), NT2-KSCN buffer containing 750 mM KSCN (twice) and PBS (once) for 5 minutes at 4°C and eluted by 2 mM D-biotin in PBS. The eluted protein complexes were denatured, reduced, alkylated and digested with immobilized trypsin (Promega) for MS analysis at MD Anderson Cancer Center Proteomics Facility.
In Vivo Breast Cancer Metastasis Assays
All animal studies were performed with MD Anderson Cancer Center's Institutional Animal Care and Use Committee (IACUC) approval. In vivo spontaneous and experimental breast cancer metastasis assays were performed as described (Chen et al., 2012; Minn et al., 2005). For animal study with LNA injection, mice were intravenously injected with in vivo grade LNAs (Exiqon) in PBS (15 mg/kg), twice a week for three weeks, after MDA-MB-231 LM2 cells injection. The tumor growth and lung metastasis were monitored by Xenogen IVIS 100 Imaging System.
Data Analysis and Statistics
Relative quantities of gene expression level were normalized to B2M. The relative quantities of ChIP and ChIRP samples were normalized by individual inputs, respectively. Results are reported as mean ± standard error of the mean (SEM) of three independent experiments. Comparisons were performed using two tailed paired Student's t test. *p < 0.05, **p < 0.01, and ***p < 0.001. Fisher exact test was used for statistical analyses of the correlation between each marker and clinical parameters. For survival analysis, the expression of BCAR4 was treated as a binary variable divided into ‘high’ and ‘low’ BCAR4 expression. Kaplan-Meier survival curves were compared by the Gehan-Breslow Test in Graphpad Prism (GraphPad Software).
Supplementary Material
Acknowledgement
We are grateful to Dr. Joan Massague and Dr. Jianming Xu for providing the MDA-MB-231 LM2 cell line and to D. Aten for assistance with figure presentation. This work was supported by NIH K99/R00 award (4R00DK094981-02), UT Startup and UT STARS grants to C.-R.L. and the NIH K99/R00 award (5R00CA166527-03), CPRIT award (R1218), UT Startup and UT STARS grants to L.-Q.Y.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
L.-Q.Y. and C.-R.L. designed the research; Z.X., A.-F.L. and C.-L.L. performed most of the biochemical and molecular experiments. A.-F.L., K.L. and L.Q. performed in vivo tumor xenograft experiments; Y.-K.W. performed pathology analyses; D.H. conducted mass spectrometry analysis. L.-Q.Y. and C.-R.L. wrote the manuscript.
References
- Carpenter S, Aiello D, Atianand MK, Ricci EP, Gandhi P, Hall LL, Byron M, Monks B, Henry-Bezy M, Lawrence JB, et al. A long noncoding RNA mediates both activation and repression of immune response genes. Science. 2013;341:789–792. doi: 10.1126/science.1240925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, Davey NE, Humphreys DT, Preiss T, Steinmetz LM, et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149:1393–1406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- Chen D, Sun Y, Wei Y, Zhang P, Rezaeian AH, Teruya-Feldstein J, Gupta S, Liang H, Lin HK, Hung MC, et al. LIFR is a breast cancer metastasis suppressor upstream of the Hippo-YAP pathway and a prognostic marker. Nat Med. 2012;18:1511–1517. doi: 10.1038/nm.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciurciu A, Duncalf L, Jonchere V, Lansdale N, Vasieva O, Glenday P, Rudenko A, Vissi E, Cobbe N, Alphey L, et al. PNUTS/PP1 regulates RNAPII-mediated gene expression and is necessary for developmental growth. PLoS genetics. 2013;9:e1003885. doi: 10.1371/journal.pgen.1003885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias N, Stein CA. Antisense oligonucleotides: basic concepts and mechanisms. Molecular cancer therapeutics. 2002;1:347–355. [PubMed] [Google Scholar]
- Dunaeva M, Michelson P, Kogerman P, Toftgard R. Characterization of the physical interaction of Gli proteins with SUFU proteins. J Biol Chem. 2003;278:5116–5122. doi: 10.1074/jbc.M209492200. [DOI] [PubMed] [Google Scholar]
- Durocher D, Taylor IA, Sarbassova D, Haire LF, Westcott SL, Jackson SP, Smerdon SJ, Yaffe MB. The molecular basis of FHA domain:phosphopeptide binding specificity and implications for phospho-dependent signaling mechanisms. Mol Cell. 2000;6:1169–1182. doi: 10.1016/s1097-2765(00)00114-3. [DOI] [PubMed] [Google Scholar]
- Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- Favoni RE, de Cupis A. The role of polypeptide growth factors in human carcinomas: new targets for a novel pharmacological approach. Pharmacological reviews. 2000;52:179–206. [PubMed] [Google Scholar]
- Feldmann G, Dhara S, Fendrich V, Bedja D, Beaty R, Mullendore M, Karikari C, Alvarez H, Iacobuzio-Donahue C, Jimeno A, et al. Blockade of hedgehog signaling inhibits pancreatic cancer invasion and metastases: a new paradigm for combination therapy in solid cancers. Cancer Res. 2007;67:2187–2196. doi: 10.1158/0008-5472.CAN-06-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii M, Lyakh LA, Bracken CP, Fukuoka J, Hayakawa M, Tsukiyama T, Soll SJ, Harris M, Rocha S, Roche KC, et al. SNIP1 is a candidate modifier of the transcriptional activity of c-Myc on E box-dependent target genes. Mol Cell. 2006;24:771–783. doi: 10.1016/j.molcel.2006.11.006. [DOI] [PubMed] [Google Scholar]
- Gai M, Camera P, Dema A, Bianchi F, Berto G, Scarpa E, Germena G, Di Cunto F. Citron kinase controls abscission through RhoA and anillin. Mol Biol Cell. 2011;22:3768–3778. doi: 10.1091/mbc.E10-12-0952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S, Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol. 2013;14:699–712. doi: 10.1038/nrm3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godinho M, Meijer D, Setyono-Han B, Dorssers LC, van Agthoven T. Characterization of BCAR4, a novel oncogene causing endocrine resistance in human breast cancer cells. J Cell Physiol. 2011;226:1741–1749. doi: 10.1002/jcp.22503. [DOI] [PubMed] [Google Scholar]
- Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui M, Cazet A, Nair R, Watkins DN, O'Toole SA, Swarbrick A. The Hedgehog signalling pathway in breast development, carcinogenesis and cancer therapy. Breast cancer research : BCR. 2013;15:203. doi: 10.1186/bcr3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakinuma T, Hwang ST. Chemokines, chemokine receptors, and cancer metastasis. Journal of leukocyte biology. 2006;79:639–651. doi: 10.1189/jlb.1105633. [DOI] [PubMed] [Google Scholar]
- Kim RH, Flanders KC, Birkey Reffey S, Anderson LA, Duckett CS, Perkins ND, Roberts AB. SNIP1 inhibits NF-kappa B signaling by competing for its binding to the C/H1 domain of CBP/p300 transcriptional co-activators. J Biol Chem. 2001;276:46297–46304. doi: 10.1074/jbc.M103819200. [DOI] [PubMed] [Google Scholar]
- Kim RH, Wang D, Tsang M, Martin J, Huff C, de Caestecker MP, Parks WT, Meng X, Lechleider RJ, Wang T, et al. A novel smad nuclear interacting protein, SNIP1, suppresses p300-dependent TGF-beta signal transduction. Genes Dev. 2000;14:1605–1616. [PMC free article] [PubMed] [Google Scholar]
- Kim YM, Watanabe T, Allen PB, Kim YM, Lee SJ, Greengard P, Nairn AC, Kwon YG. PNUTS, a protein phosphatase 1 (PP1) nuclear targeting subunit. Characterization of its PP1- and RNA-binding domains and regulation by phosphorylation. J Biol Chem. 2003;278:13819–13828. doi: 10.1074/jbc.M209621200. [DOI] [PubMed] [Google Scholar]
- Komarnitsky P, Cho EJ, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 2000;14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling H, Fabbri M, Calin GA. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nature reviews Drug discovery. 2013;12:847–865. doi: 10.1038/nrd4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunde BM, Moore C, Varani G. RNA-binding proteins: modular design for efficient function. Nat Rev Mol Cell Biol. 2007;8:479–490. doi: 10.1038/nrm2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madaule P, Eda M, Watanabe N, Fujisawa K, Matsuoka T, Bito H, Ishizaki T, Narumiya S. Role of citron kinase as a target of the small GTPase Rho in cytokinesis. Nature. 1998;394:491–494. doi: 10.1038/28873. [DOI] [PubMed] [Google Scholar]
- Maloverjan A, Piirsoo M, Michelson P, Kogerman P, Osterlund T. Identification of a novel serine/threonine kinase ULK3 as a positive regulator of Hedgehog pathway. Exp Cell Res. 2010;316:627–637. doi: 10.1016/j.yexcr.2009.10.018. [DOI] [PubMed] [Google Scholar]
- Meijer D, van Agthoven T, Bosma PT, Nooter K, Dorssers LC. Functional screen for genes responsible for tamoxifen resistance in human breast cancer cells. Molecular cancer research : MCR. 2006;4:379–386. doi: 10.1158/1541-7786.MCR-05-0156. [DOI] [PubMed] [Google Scholar]
- Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, Viale A, Olshen AB, Gerald WL, Massague J. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller A, Homey B, Soto H, Ge N, Catron D, Buchanan ME, McClanahan T, Murphy E, Yuan W, Wagner SN, et al. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410:50–56. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin LL, de Sauvage FJ. Targeting the Hedgehog pathway in cancer. Nature reviews Drug discovery. 2006;5:1026–1033. doi: 10.1038/nrd2086. [DOI] [PubMed] [Google Scholar]
- ten Haaf A, Bektas N, von Serenyi S, Losen I, Arweiler EC, Hartmann A, Knuchel R, Dahl E. Expression of the glioma-associated oncogene homolog (GLI) 1 in human breast cancer is associated with unfavourable overall survival. BMC Cancer. 2009;9:298. doi: 10.1186/1471-2407-9-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelie A, Papillier P, Pennetier S, Perreau C, Traverso JM, Uzbekova S, Mermillod P, Joly C, Humblot P, Dalbies-Tran R. Differential regulation of abundance and deadenylation of maternal transcripts during bovine oocyte maturation in vitro and in vivo. BMC developmental biology. 2007;7:125. doi: 10.1186/1471-213X-7-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varjosalo M, Taipale J. Hedgehog: functions and mechanisms. Genes Dev. 2008;22:2454–2472. doi: 10.1101/gad.1693608. [DOI] [PubMed] [Google Scholar]
- Wang X, Venugopal C, Manoranjan B, McFarlane N, O'Farrell E, Nolte S, Gunnarsson T, Hollenberg R, Kwiecien J, Northcott P, et al. Sonic hedgehog regulates Bmi1 in human medulloblastoma brain tumor-initiating cells. Oncogene. 2012;31:187–199. doi: 10.1038/onc.2011.232. [DOI] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washington K, Ammosova T, Beullens M, Jerebtsova M, Kumar A, Bollen M, Nekhai S. Protein phosphatase-1 dephosphorylates the C-terminal domain of RNA polymerase-II. J Biol Chem. 2002;277:40442–40448. doi: 10.1074/jbc.M205687200. [DOI] [PubMed] [Google Scholar]
- Yamashiro S, Totsukawa G, Yamakita Y, Sasaki Y, Madaule P, Ishizaki T, Narumiya S, Matsumura F. Citron kinase, a Rho-dependent kinase, induces di-phosphorylation of regulatory light chain of myosin II. Mol Biol Cell. 2003;14:1745–1756. doi: 10.1091/mbc.E02-07-0427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Lin C, Jin C, Yang JC, Tanasa B, Li W, Merkurjev D, Ohgi KA, Meng D, Zhang J, et al. lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature. 2013;500:598–602. doi: 10.1038/nature12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, Bi L, Zheng B, Ji L, Chevalier D, Agarwal M, Ramachandran V, Li W, Lagrange T, Walker JC, et al. The FHA domain proteins DAWDLE in Arabidopsis and SNIP1 in humans act in small RNA biogenesis. Proc Natl Acad Sci U S A. 2008;105:10073–10078. doi: 10.1073/pnas.0804218105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.