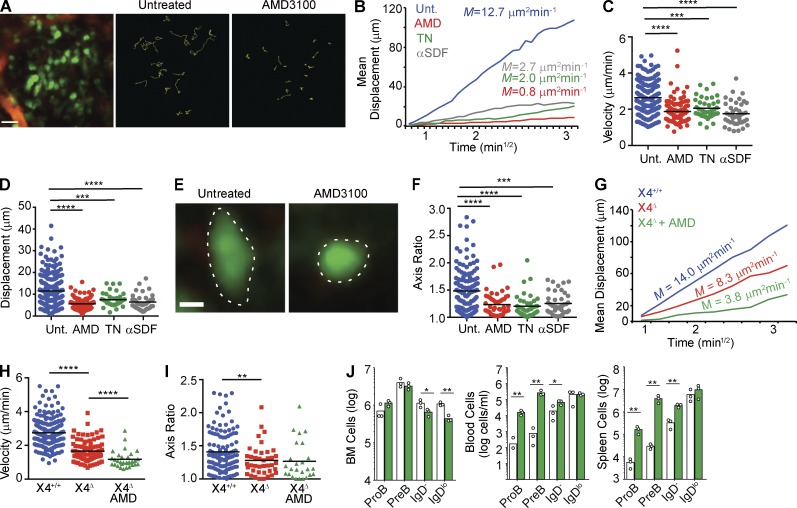
Figure 1.
CXCR4 antagonism inhibits B lineage cell migration in BM. (A–F) Rag1GFP/+ mice were injected i.v. with AMD3100, TN14003, or anti-CXCL12 antibody. Blood vessels were labeled with 2,000-kD dextran-rhodamine injected i.v. Cell movement was tracked by IVM of calvaria immediately before and after treatment. (A, left) Distribution of GFP+ B cells (green) in BM. (middle and right) Movement of GFP+ B cells tracked for 30 min before and after AMD3100 treatment, respectively. Colored lines represent cell trajectories. Data are representative of three individual experiments. (B) Mean motility coefficient of B lineage cells before (blue) and after (red) treatment with CXCR4 antagonists. Cell displacement from starting coordinates is plotted against the square root of time. Lines depict the average mean motility coefficient calculated from three individual experiments. (C) Median velocity (µm/min). (D) Displacement (µm). (E) Morphology of GFP+ B lineage cells before (left) and after (right) AMD3100 treatment. Dotted lines depict the cells’ borders. Bars: (A) 20 µm; (E) 30 µm. (F) Measurement of cell axis ratio of GFP+ B lineage cells before (blue) and after (red) treatment with CXCR4 inhibitors. (G–I) WT (Mb1Cre/+) and CXCR4 KO (Mb1Cre/+ Cxcr4Fl/−) were treated as in A–F. (G) Mean motility coefficient of WT (blue) and CXCR4 KO (red) B lineage cells before (red) and after (green) treatment with AMD3100. Colored lines depict average mean motility coefficient calculated from three independent datasets. (H) Median cell velocity of WT (blue) and CXCR4 KO (red) B lineage cells before (red) and after (green) treatment with AMD3100. (I) Cell axis ratio of GFP+ (Mb1Cre/+, blue) and CXCR4 KO (Mb1Cre/+ Cxcr4Fl/−, red) B lineage cells. (G–I) Data are representative of two independent experiments. (C, D, F, H, and I) Lines indicate mean. (J) Enumeration of B lineage cells in BM, blood, and spleen of Mb1Cre/+ Cxcr4+/+ (open bars) and Mb1Cre/+ Cxcr4Fl/− (green bars) mice. Bars indicate mean, and circles indicate individual mice. Data are representative of three independent experiments. *, P < 0.05; **, P < 0.005; ***, P < 0.0005; ****, P < 0.00005 by unpaired Student’s t test.