Figure 1.
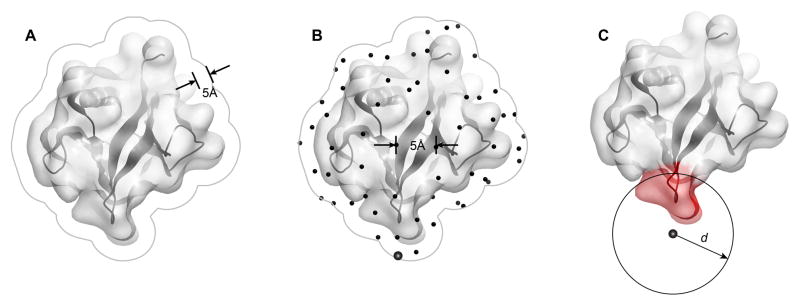
Protein surface patch generation procedure used by MODA, as illustrated using the PH domain of human B lymphocyte adapter protein Bam32 (PDB 1 fao). The generation of surface patches involves (A) expanding the solvent accessible surface by 5 Å to represent the overall protein shape rather than fine atomic details and to avoid leakage through thin sections of protein, (B) generating an even distribution of points across the surface from which to predict the membrane binding propensity, and (C) assigning each point to a patch consisting of all the proximal solvent accessible atoms within distance d Å from the point, where d and the learning algorithm are as described for the PIER program (Kufareva et al., 2007) using patch predictors which were defined from the known membrane interaction sites.
