Significance
By comparing the form of the chlorophyll fluorescence decays in wild-type, npq1, npq4, and npq1 npq4 plants, we show that the presence of violaxanthin deepoxidase (VDE), but not the protein PsbS, changes the excited-state relaxation dynamics of chlorophyll. PsbS has the strongest effect during the initial induction of quenching. The presence of both zeaxanthin and PsbS is required for strong quenching. This fluorescence lifetime technique helps elucidate both the kinetics with which quenching turns on and off and the relaxation dynamics of excited-state chlorophyll.
Keywords: PsbS, nonphotochemical quenching, fluorescence lifetime, carotenoids, photosystem II
Abstract
The photosystem II (PSII) protein PsbS and the enzyme violaxanthin deepoxidase (VDE) are known to influence the dynamics of energy-dependent quenching (qE), the component of nonphotochemical quenching (NPQ) that allows plants to respond to fast fluctuations in light intensity. Although the absence of PsbS and VDE has been shown to change the amount of quenching, there have not been any measurements that can detect whether the presence of these proteins alters the type of quenching that occurs. The chlorophyll fluorescence lifetime probes the excited-state chlorophyll relaxation dynamics and can be used to determine the amount of quenching as well as whether two different genotypes with the same amount of NPQ have similar dynamics of excited-state chlorophyll relaxation. We measured the fluorescence lifetimes on whole leaves of Arabidopsis thaliana throughout the induction and relaxation of NPQ for wild type and the qE mutants, npq4, which lacks PsbS; npq1, which lacks VDE and cannot convert violaxanthin to zeaxanthin; and npq1 npq4, which lacks both VDE and PsbS. These measurements show that although PsbS changes the amount of quenching and the rate at which quenching turns on, it does not affect the relaxation dynamics of excited chlorophyll during quenching. In addition, the data suggest that PsbS responds not only to ΔpH but also to the Δψ across the thylakoid membrane. In contrast, the presence of VDE, which is necessary for the accumulation of zeaxanthin, affects the excited-state chlorophyll relaxation dynamics.
Plants regulate light harvesting by photosystem II (PSII) in response to changes in light intensity. One way that plants are able to regulate light harvesting is through turning on and off mechanisms that dissipate excess energy. This energy dissipation is assessed via nonphotochemical quenching (NPQ) measurements of chlorophyll fluorescence. Energy-dependent quenching (qE) is the NPQ process with the fastest kinetics. It turns on and off in seconds to minutes, allowing plants to respond to rapid fluctuations in light intensity, which is thought to reduce photodamage (1, 2).
Illumination causes the formation of gradients of electrical potential (Δψ) and of proton concentration (ΔpH) across the thylakoid membrane. Although it has been suggested that Δψ may play a role in qE (3), only ΔpH is thought to trigger different proteins and enzymes to induce qE (4). The major known factors involved in induction of qE are the enzyme violaxanthin deepoxidase (VDE) (5) and the PSII protein PsbS (6). The mutant npq1, which lacks VDE and cannot convert violaxanthin to zeaxanthin, has a phenotype with lower qE compared with the wild type (7). Transient absorption measurements suggest that zeaxanthin may quench excited chlorophyll (8). The npq4 mutant, which lacks PsbS, shows no rapidly reversible quenching of chlorophyll fluorescence, suggesting that PsbS is required for qE in vivo (6). PsbS is pH sensitive (9) but is not thought to bind pigments, and thus is likely not the site of quenching (10). It has therefore been hypothesized that PsbS plays an indirect role in quenching, perhaps facilitating a rearrangement of proteins within the grana (11–13). In this paper, we examine the fluorescence lifetime of chlorophyll throughout the induction and relaxation of quenching in intact leaves with and without PsbS and zeaxanthin to examine whether PsbS and zeaxanthin change the type of quenching that occurs in plants.
The amount and dynamics of qE are generally measured by changes in the chlorophyll fluorescence yield. One limitation of the chlorophyll fluorescence yield is that it can only inform on the amount of quenching, not on excited-state chlorophyll relaxation dynamics, which reflect how chlorophyll is quenched. Despite this issue, the amount of quenching is commonly used as a proxy for the type of quenching by separating components of quenching based on kinetics, mutants, and the effects of chemical inhibitors. By artificially increasing ΔpH in isolated chloroplasts from npq4, Johnson and Ruban (14, 15) have been able to increase the amount of quenching in npq4 plants to levels observed in wild type plants, suggesting that PsbS may catalyze qE. One potential complication with these studies is that the use of the chemical mediators of cyclic electron transport often necessitates studying isolated chloroplasts rather than intact leaves. In addition, the observation of equivalent amounts of quenching still does not prove that the type of quenching in npq4 is the same as in wild type.
In contrast with fluorescence yield measurements, fluorescence lifetime measurements can be used to determine whether the relaxation dynamics of excited chlorophyll are modified by different mutations, informing on the role of a protein or molecule during quenching. The relaxation dynamics of excited chlorophyll during NPQ depends on many variables, including the distance to a quencher, the interactions between the orbitals of chlorophyll and the quencher, and the number of quenchers (16). The shape of the fluorescence lifetime decay curve can be used to determine whether two samples have similar excited chlorophyll relaxation dynamics. Our results show that, although the presence of PsbS does not alter excited chlorophyll relaxation dynamics, the absence of VDE does. These measurements are performed in intact leaves without any chemical treatments, and the data strongly suggest that PsbS plays a catalytic role in vivo.
Results
To examine the dynamics of quenching, fluorescence lifetimes were measured for wild-type, npq4, npq1, and npq1 npq4 leaves during a 45-min illumination period with 500 μmol photons⋅m−2⋅s−1 light. To deconvolute the dynamics of qE from NPQ mechanisms that relax on a longer timescale, the actinic light was subsequently turned off for 3 min. This amount of time is long enough to dissipate the ΔpH that triggers qE (17), but not long enough for significant conversion of zeaxanthin back to violaxanthin, which is necessary to turn off a zeaxanthin-dependent but ΔpH-independent component of NPQ called qZ (18). The actinic light was then turned on for a 10-min period to turn qE back on.
Amplitude-Weighted Average Fluorescence Lifetimes.
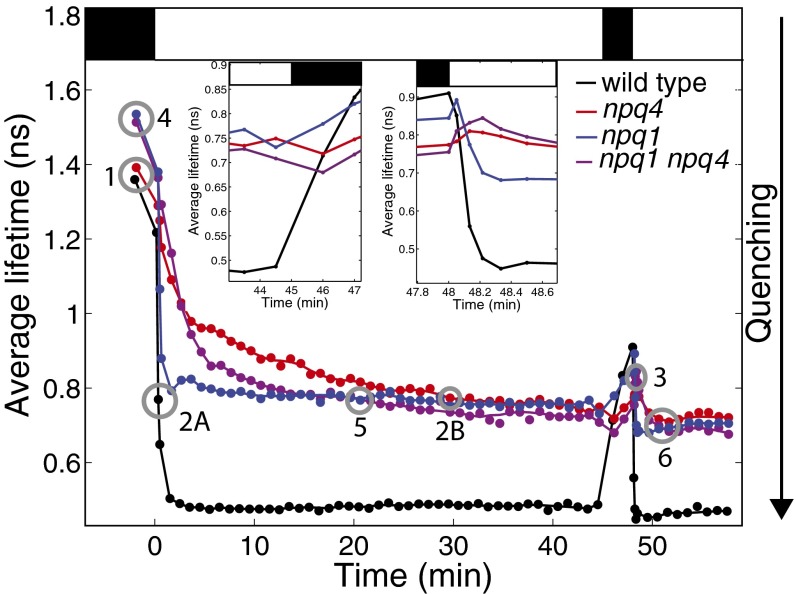
The amplitude-weighted average fluorescence lifetimes for wild type, npq4, npq1, and npq1 npq4 over the duration of the experiments are shown in Fig. 1. The light sequence of the actinic light is shown by the white and black bars at the Top of Fig. 1. Both wild-type and npq4 leaves had nearly equal average fluorescence lifetimes in the dark. Both zeaxanthin-free mutants (npq1 and npq1 npq4) had nearly equal dark-adapted fluorescence lifetimes (though the lifetimes were longer than for wild type and npq4). When the actinic light was turned on, the average fluorescence lifetime decreased for all of the genotypes. However, the fluorescence lifetime for plants with PsbS (wild type and npq1) decreased more rapidly than the mutants without PsbS (npq4 and npq1 npq4). After 30 min of illumination, npq4, npq1, and npq1 npq4 all reached approximately the same average fluorescence lifetime of 0.75 ns, whereas wild type had an average lifetime of 0.47 ns.
Fig. 1.
Average fluorescence lifetimes of wild type (black), npq1 (blue), npq4 (red), and npq1 npq4 (purple) are shown as closed circles. The gray open circles indicate the two similar average fluorescence lifetimes that are used to compare the shapes of the fluorescence lifetime decays. Circles 1, 2A and 2B, and 3 indicate comparisons between wild-type and npq4 dark-adapted leaves and during the first and second illumination periods, respectively. Circles 4, 5, and 6 correspond to comparisons between npq1 and npq1 npq4 for dark-adapted leaves, and during the first and second illumination periods, respectively. (Top) Actinic light on (white bar) and off (black bar). (Insets) Zoomed-in portions of the average fluorescence lifetimes. Leaves were illuminated with 500 μmol photons⋅m−2⋅s−1 actinic light.
One minute after the actinic light was turned off, the leaves that contain PsbS (wild type and npq1) exhibited an increase in average fluorescence lifetime. In contrast, both npq4 and npq1 npq4 showed a transient decrease in the average fluorescence lifetime, dropping by ∼30 ps (Left Inset of Fig. 1). After this drop, the average fluorescence lifetime increased over the next 2 min of darkness. When the actinic light was turned on for the second time, the average fluorescence lifetime of wild type decreased by 40 ps within 3 s of illumination, whereas the fluorescence lifetimes of all of the mutants increased. The average fluorescence lifetime of npq1, npq4, and npq1 npq4 dropped after 8, 13, and 20 s of illumination, respectively (Right Inset of Fig. 1). Again, the fluorescence lifetimes for wild type and npq1 decrease more rapidly than those of npq4 and npq1 npq4.
Differences in the Average Fluorescence Lifetimes Between Pairs of Plants.
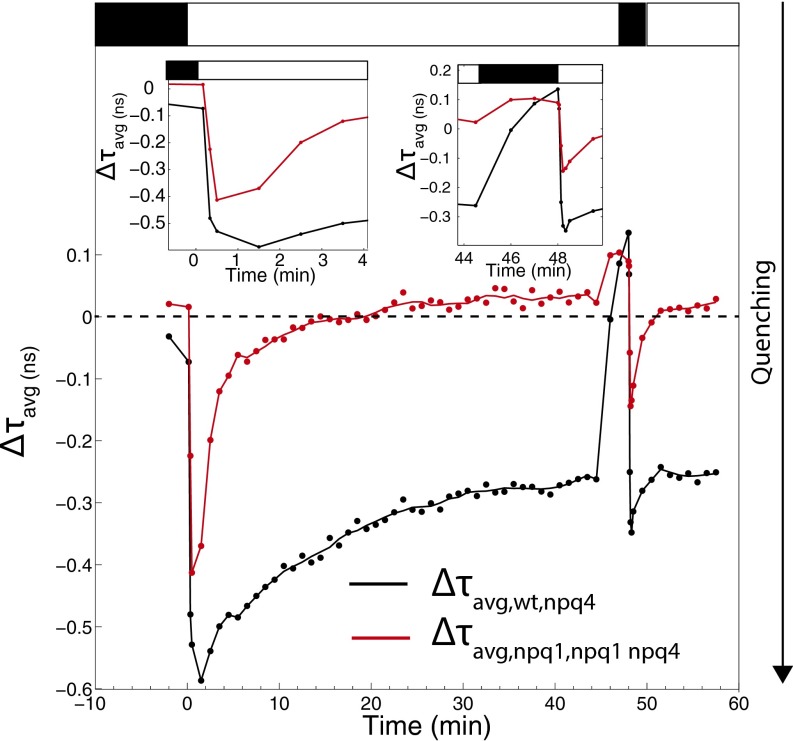
The difference between the average fluorescence lifetimes, can give insight into how different components influence the dynamics of NPQ. Both and are shown in Fig. 2. This figure shows the effect of PsbS on quenching in the presence and absence of zeaxanthin.
Fig. 2.
The effect of PsbS on the amount of quenching. (black) and (red) are shown. is calculated by subtracting the average fluorescence lifetime of npq4 from that of wild type, and is calculated by subtracting the average fluorescence lifetime of npq1 npq4 from that of npq1. When the value of or equals 0 (shown by the dashed line), it indicates no difference in average lifetime between the two genotypes. A negative value of means that wild type has a shorter average fluorescence lifetime, i.e., stronger quenching, than npq4, and a negative value of means that npq1 has stronger quenching than npq1 npq4. (Insets) Zoomed-in portions of the and . Leaves were illuminated with 500 μmol photons⋅m−2⋅s−1 actinic light.
The absolute value of increased over the first 1.5 min of illumination, with the biggest difference occurring between 10 and 30 s of illumination (Left Inset of Fig. 2). After 1.5 min of illumination, the absolute value of decreased as the induction of quenching in wild type slowed down and reached its steady-state level, whereas npq4 continued to experience an increase in quenching. These dynamics suggest that PsbS plays a major role primarily within the first minute of illumination (Right Inset of Fig. 2). During the second illumination period, showed similar dynamics to what was observed during the first illumination period, with the biggest change seen between 3 and 8 s of the second illumination. After 2.5 min of the second illumination, the difference between wild type and npq4 stabilized.
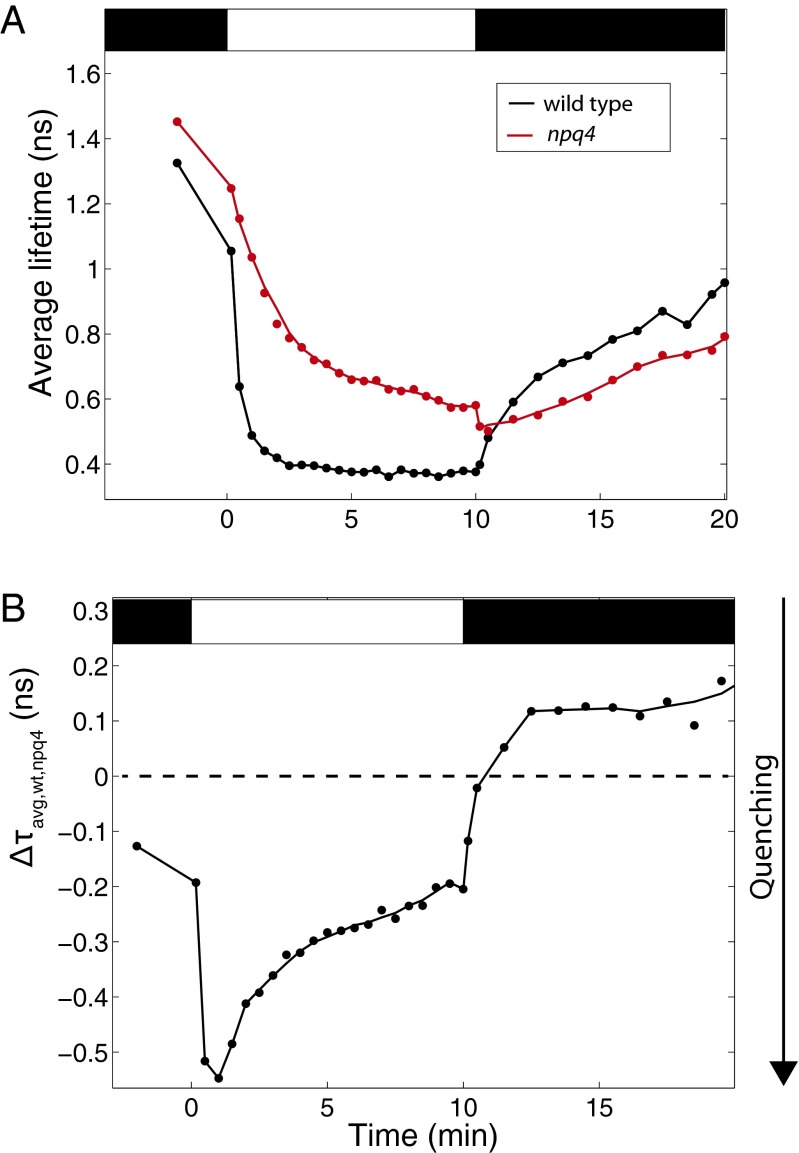
To further examine the PsbS-dependent relaxation kinetics, wild-type and npq4 plants were illuminated for 10 min with higher light intensity (1,200 μmol photons⋅m−2⋅s−1) and then allowed to relax for 10 min. The average fluorescence lifetimes are shown in Fig. 3A, and the difference in average fluorescence lifetime is shown in Fig. 3B. With this higher actinic light intensity, both wild type and npq4 reached shorter lifetimes than when illuminated with 500 μmol photons⋅m−2⋅s−1 light, but the average lifetime of npq4 was closer to that of wild type after 10 min of illumination. When the actinic was light turned off, wild type experienced a biphasic decrease in quenching, but npq4 had a sudden increase in quenching during the first 30 s of darkness, followed by a linear decrease in quenching. After 2 min of darkness, wild type and npq4 recovered at the same rate. This result shows that, during recovery in the dark, as during illumination, PsbS only affects the quenching dynamics in the first minutes following the transition between light and dark.
Fig. 3.
The effect of PsbS during high-light illumination. The average fluorescence lifetimes of wild type (black) and npq4 (red) during 10 min of illumination with 1,200 μmol photons⋅m−2⋅s−1 and 10 min of relaxation are shown in A. The difference between average fluorescence lifetimes of wild type and npq4, , is shown in B.
To examine whether the presence of zeaxanthin changes the role of PsbS, the difference in average fluorescence lifetime was also calculated for npq1 and npq1 npq4 (Fig. 2, red trace). Similar to , during the first minutes of light, the dynamics of had a negative peak following the transition from dark to light. However, without zeaxanthin, the largest magnitude of shifted 30 s earlier, occurring after 1 min of illumination. The magnitude of the lifetime difference was also slightly smaller. In addition, the decrease in the magnitude of happened more quickly compared with . When the actinic light was turned off, reached a steady state within a minute, indicating that both npq1 and npq1 npq4 relaxed at the same rate.
Fluorescence Decay Comparisons Between Wild Type and npq4.
To examine whether the presence of PsbS impacts the relaxation dynamics of excited chlorophyll, the shapes of the fluorescence lifetime decays were compared between wild type and npq4 at three different sets of points: on dark-adapted leaves before exposure to actinic light, on leaves with quenching on during the first illumination period, and on leaves with quenching on during the second illumination period. These comparison points are shown by gray circles 1, 2A and 2B, and 3 in Fig. 1.
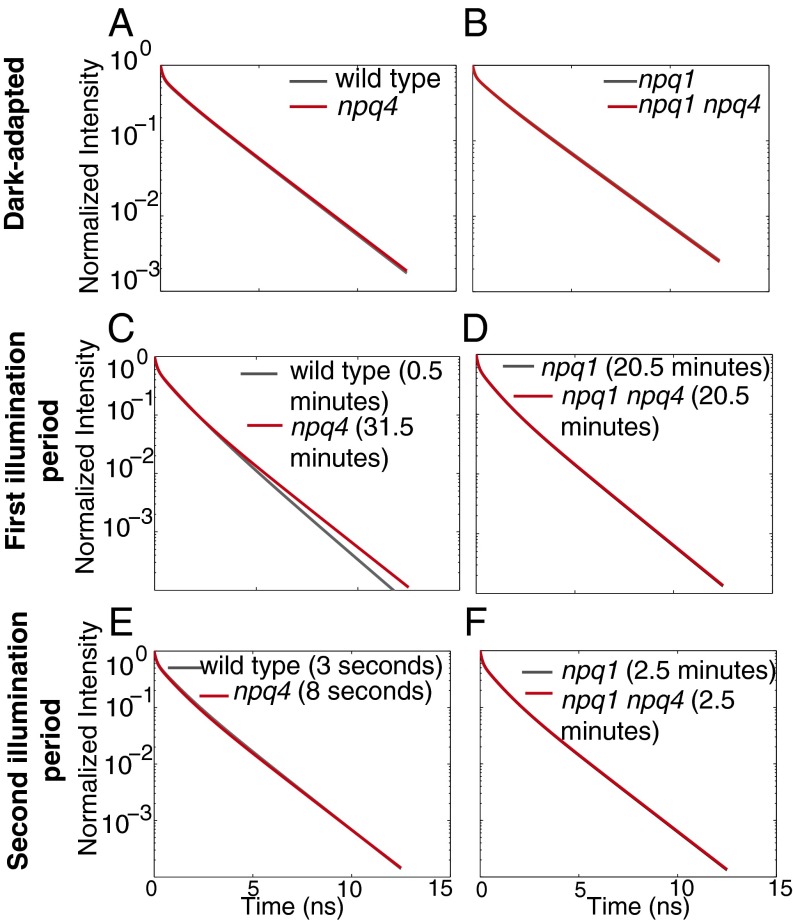
The comparison between dark-adapted wild type and npq4 leaves shows whether the presence of PsbS impacts the relaxation of excited chlorophyll before light treatment. The fluorescence decays, reconstructed by the fit to the data, are shown in Fig. 4A. In dark-adapted leaves, the shapes of the fluorescence decays were identical for wild type and npq4. This is confirmed by examining the values for the fluorescence lifetime components and their associated amplitudes, shown in Fig. S1 A and B.
Fig. 4.
Comparisons of the fluorescence decay between wild type (Left, gray) and npq4 (Left, red) and between npq1 (Right, gray) and npq1 npq4 (Right, red) for different values of the average fluorescence lifetime. The fluorescence lifetime decays of wild type and npq4 in the dark-adapted state (A), after 30 s and 31.5 min of illumination for wild type and npq4, respectively (C), and during the second illumination period after 45 min of first illumination period followed by 3 min in the dark, 3 s and 8 s of illumination for wild type and npq4, respectively (E). The fluorescence lifetime decays of npq1 and npq1 npq4 in the dark-adapted state (B), after 20.5 min of illumination (D), and during the second illumination period after 2.5 min of illumination (F). Leaves were illuminated with 500 μmol photons⋅m−2⋅s−1 actinic light.
Upon induction of NPQ, both genotypes reached an average fluorescence lifetime of 0.77 ns during illumination (0.77 ± 0.03 ns for wild type after 30 s of illumination and 0.77 ± 0.02 ns for npq4 after 31.5 min of illumination). This average fluorescence lifetime corresponds to an NPQ value of 0.8, compared with a maximum NPQ value of 1.9 for wild type and 0.9 for npq4 (see SI Text for derivation of the calculation). The shapes of the fluorescence decays are shown in Fig. 4C. Despite equal values of average fluorescence lifetime, the fluorescence decays shapes were different for the two genotypes. The long decay component was shorter for wild type compared with npq4 and the medium component was longer for wild type compared with npq4, as shown in Fig. S1C. The amplitudes for each of the lifetime components were similar for wild type and npq4, as shown in Fig. S1D. The parameters that describe the fluorescence lifetime decays for wild type and npq4 differ by a statistically significant amount, and we were unable to find a set of parameters that was able to describe both genotypes. This difference indicates different excited-state chlorophyll relaxation dynamics for wild type and npq4.
To compare wild type and npq4 with equal fluorescence lifetimes during quenching, it was necessary to compare the plants when they had experienced different illumination durations. It is known that the concentration of zeaxanthin changes during illumination on a timescale of minutes to tens of minutes (18). Therefore, we hypothesized that the difference in fluorescence decay shapes was due to a difference in zeaxanthin concentration, which would affect the amount of qZ. When wild-type and npq4 leaves were illuminated with 500 μmol photons⋅m−2⋅s−1 actinic light for 30 s and 30 min, there was no zeaxanthin detected in either wild type or npq4 after 30 s of illumination (Table S1). However, after 30 min of illumination, there was ∼21 mmol of zeaxanthin per mol of chlorophyll a detected in wild type, suggesting that the difference in fluorescence lifetime decay shape is due to a difference in qZ. To test whether the presence of PsbS alters the chlorophyll relaxation dynamics when there is zeaxanthin present in both wild type and npq4, the shape of the fluorescence lifetime decays was compared during the second illumination period. Both wild-type and npq4 leaves reached equivalent values for the average fluorescence lifetime after 3 and 8 s of illumination, respectively. The fluorescence decays are shown in Fig. 4E. The fitting parameters are shown in Fig. S1 E and F. In this comparison, the shape of the fluorescence lifetime decays of wild type and npq4 were identical.
Fluorescence Decay Comparisons Between npq1 and npq1 npq4.
If the difference in quenching shown in Fig. 4C was due to a difference in the zeaxanthin concentration (and amount of qZ), it is expected that without zeaxanthin present, the presence of PsbS will not change the relaxation dynamics of excited chlorophyll during quenching. To test this prediction, fluorescence lifetimes were measured on npq1 and npq1 npq4. Due to the similarity of the average fluorescence lifetime traces between these two mutants, there were multiple points at which the average fluorescence lifetime curves for npq1 and npq1 npq4 crossed. This allows for comparisons under equivalent illumination conditions. We were able to make three comparisons: dark-adapted npq1 compared with dark-adapted npq1 npq4, npq1 and npq1 npq4 after 20.5 min of illumination, and npq1 and npq1 npq4 after 2.5 min of illumination in the second illumination period. These comparison points are shown by circles 4, 5, and 6 in Fig. 1.
The comparison between dark-acclimated npq1 and npq1 npq4 is shown in Fig. 4B. The fluorescence decay shapes were identical. The fitting parameters, shown in Fig. S2 A and B, were also identical within error. This result confirms that the presence of PsbS does not affect the relaxation dynamics of chlorophyll for dark-adapted leaves. Next, the fluorescence lifetimes of light-adapted npq1 and npq1 npq4 were compared to examine how the presence of PsbS affects the dynamics of quenching when NPQ is on, but there is no zeaxanthin present. The fluorescence decays were compared where the average fluorescence lifetime curves cross at 20.5 min of illumination at an average fluorescence lifetime of 0.77 ns. The fluorescence decays for these two mutants are shown in Fig. 4D. Again, the decay curves were identical. This can also be seen in the fluorescence decay fit parameters shown in Fig. S2 C and D. Last, the fluorescence decays for npq1 and npq1 npq4 were compared during the second illumination time after 2.5 min of illumination. At this time, the average fluorescence lifetimes were 0.69 ns for npq1 npq4 and 0.70 ns for npq1 npq4. The fluorescence decays for this comparison are shown in Fig. 4F and are very similar. The fitting parameters are shown in Fig. S2 E and F. In all three sets of comparisons, the fluorescence decays for both npq1 and npq1 npq4 were essentially identical, despite the difference in the presence or absence of PsbS. This indicates that the presence of PsbS does not significantly impact the quenching pathways for excited chlorophyll in the absence of zeaxanthin, regardless of the amount of quenching.
Discussion
We measured the fluorescence lifetimes on whole leaves of wild-type, npq1, npq4, and npq1 npq4 plants as they acclimate to changing light conditions. Examining the dynamics of the average fluorescence lifetimes reveals information about how PsbS and zeaxanthin influence the dynamics of qE as it turns on and off. The fluorescence decay similarity between plants with and without PsbS can be seen when quenching is off (Fig. 4 A and B) and when quenching is on (Fig. 4 D–F). The difference in the form of the fluorescence decay between wild-type leaves that were illuminated for 30 s and for npq4 leaves that were illuminated for 31.5 min (shown in Fig. 4C) is most likely due to a difference in the contribution of the zeaxanthin-dependent component of NPQ. When zeaxanthin was present in equivalent amounts (shown in Fig. 4E) or when zeaxanthin was absent, the fluorescence lifetime decays for plants with and without PsbS were identical. This evidence supports the hypothesis that PsbS acts as a catalyst to accelerate the induction of quenching, but does not affect the possible relaxation pathways.
The ability to convert violaxanthin to zeaxanthin does appear to strongly impact the dynamics of chlorophyll relaxation. Fig. 1 shows that dark-adapted npq1 and npq1 npq4 leaves have a longer average fluorescence lifetime than leaves that contain VDE. Although it should be noted that the dark-adapted fluorescence lifetime of plants seems to vary based on growth conditions (as can be seen by comparing the average fluorescence lifetimes of plants in Figs. 1 and 3), the difference between plants with and without VDE is reproducible between batches of plants grown in different growth chambers at different times. Although our HPLC measurements were unable to detect zeaxanthin in dark-adapted wild-type plants (Table S1), there is likely a small amount of zeaxanthin present (7) and we detected minor amounts of antheraxanthin, which is linked to energy dissipation (19). The fluorescence lifetime comparisons between wild type and npq4 (Fig. 4 A, C, and E) show that the excited chlorophyll relaxation dynamics differ only when the concentration of zeaxanthin is different. The strong effect of zeaxanthin could be due to zeaxanthin directly acting as a quencher (8) or by influencing the thylakoid membrane structure (20). Because zeaxanthin changes the relaxation dynamics of excited chlorophyll in addition to the amount of quenching, taking the difference in average fluorescence lifetime between wild type and npq1 or between npq4 and npq1 npq4 cannot clearly show the effect of zeaxanthin.
The large negative peak in the difference in average fluorescence lifetimes, and in Figs. 2 and 3B, show that the presence of PsbS is most important during the first minute of illumination. After 1 min of illumination, the role of PsbS is less marked. In the second illumination period, this negative peak appeared earlier, occurring within 30 s of illumination. The timing of the negative peak is surprising, because it suggests that the presence of PsbS is most important during the increase of the pH gradient rather than when the ΔpH reaches its steady-state value, which occurs after 1–1.5 min of illumination (15, 21), depending on the illumination intensity and on the ion concentration (22). When plants are exposed to actinic light, a proton motive force forms, first in the form of Δψ, which then dissipates as ΔpH increases (22). After 20.5 min of exposure to the actinic light, reaches a value of zero, indicating that, at this point, the presence of PsbS no longer increased the amount of quenching, despite the maintenance of a pH gradient. The different dynamics of and indicate that the presence of zeaxanthin influences the role of PsbS. The fact that npq1, npq4, and npq1 npq4 all reach the same minimum value of average fluorescence lifetime after 45 min of illumination, as shown in Fig. 1, suggests that zeaxanthin and PsbS interact in wild-type leaves to cause a full quenching response, and the loss of either or both of these components has the same effect on the maximum amount of quenching observed. These results, along with the altered relaxation dynamics of chlorophyll when VDE is absent, suggest that the kinetics of qZ cannot easily be disentangled from that of qE due to the interaction between PsbS and zeaxanthin.
When the actinic light is turned off, Δψ decreases to negative values and ΔpH decreases toward zero as protons move from the lumen to the stroma (22). Without PsbS, quenching increased when the actinic light was turned off (Figs. 1 and 3A). This increase would not be expected if quenching was only sensitive to the ΔpH, because during this time period, ΔpH is decreasing toward zero (15, 21) and a lowered ΔpH corresponds to a lower amount of quenching (23). However, during this time period, the electric potential gradient, Δψ, reaches negative values, which does not occur at any other times during quenching (21, 22). This finding suggests that the increase in quenching could be due to Δψ.
Further evidence that Δψ may influence the quenching dynamics can be seen in the plots of shown in Fig. 3B. The shape of resembles the dynamics of −Δψ during the induction of quenching, but resembles the dynamics of ΔpH during the relaxation of quenching (21, 22). Because shows the difference in quenching due to PsbS, this suggests that PsbS may respond to Δψ in addition to ΔpH. Although studies have confirmed that mutating protonatable residues in PsbS results in the same phenotype as npq4 (9), protonatable residues are likely also responsive to positive ions other than protons. Therefore, any situation that would disrupt the protonation of PsbS would also destroy its ability to respond to other ions. We therefore hypothesize that, initially, the interaction between PsbS and an excess of positive cations in the lumen causes the switch of PsbS to a quenching-inducing state. As Δψ decreases and ΔpH increases, the interaction between PsbS and nonproton cations such as K+ is replaced by the increasing number of protons. When the actinic light is turned off, ΔpH decreases and Δψ becomes negative as protons move from the lumen to the stroma (24). The resemblance of to the ΔpH during the relaxation of quenching can therefore easily be explained by the fact that the ions that are moving are protons rather than other positive ions. When the actinic light is turned off, the increase in quenching when PsbS is absent and the fast decrease in quenching when PsbS is present, suggests that the decreases in ΔpH and Δψ cause PsbS to switch to a state that facilitates the relaxation of quenching.
In summary, our results show that, although PsbS does not change the relaxation pathways available to chlorophyll, it serves three important roles. (i) It enables plants to sense changes in light intensity in under 30 s. (ii) It protects plants from experiencing an increase in quenching when the light turns off. (iii) PsbS, in concert with zeaxanthin, allows plants to reach a configuration in which many more chlorophylls have access to NPQ relaxation pathways. These results suggest that PsbS not only helps the plant respond quickly to changes in light intensity, but switches quenching on and off before the photosynthetic machinery would be able to do so otherwise. Furthermore, our results suggest that the presence of zeaxanthin allows plants with PsbS to rapidly attain the strongest quenching. This interaction between PsbS and zeaxanthin may help plants under natural fluctuating light conditions rapidly respond to excess light. In the future, the technique of comparing fluorescence lifetimes can be used to study other NPQ components and the interactions between proteins and processes involved, such as the interaction between state transitions and PsbS, or the interaction between fast-timescale quenching processes and longer processes such as photoinhibition.
Conclusions
The fluorescence lifetime measurements of wild type, npq4, npq1, and npq1 npq4 plants during induction and relaxation of NPQ give insight into how PsbS and zeaxanthin affect the amount and type of quenching. We have found that PsbS is responsible for the initial quenching processes, with its action occurring most strongly in the first minute of illumination and in the first 2 min of relaxation. The absence of either zeaxanthin, PsbS, or the absence of both at the same time gives the same amount of quenching as measured by the average fluorescence lifetime after 45 min of illumination. This result suggests that the high amount of quenching seen in wild type requires the presence of both zeaxanthin and PsbS, and that it is due to an interaction between PsbS and zeaxanthin. Although PsbS changes the amount of possible quenching, and the rate at which quenching turns on, it does not affect the excited-state relaxation dynamics of chlorophyll. In contrast, the presence of zeaxanthin changes both the timescale over which quenching turns on and off, and the relaxation dynamics of excited chlorophyll.
Materials and Methods
Plant Material and Growth Conditions.
Arabidopsis thaliana wild-type (ecotype Columbia 0) and mutant plants npq1 (7), npq4 (6), and npq1 npq4 (25) were grown on soil at a light intensity of 150 μmol photons⋅m−2⋅s−1 under short day conditions (8 h light, 22 °C/16 h dark, 23 °C). Six- to 9-wk-old plants were used for all experiments. Replicates were done using leaves from different plants grown at different times. The plants were each dark-adapted for 30 min before detaching a leaf.
Fluorescence Lifetime Measurement.
Experiments were conducted using a home-built fluorescence lifetime measurement apparatus. Similar to the setup described by Amarnath et al. (26), the light exposure of the leaf and detector were controlled by shutters programmed in LabVIEW. A 532-nm diode laser (Coherent G10) was used to pump a Ti:sapphire oscillator (Coherent Mira 900) set to 840 nm. The resulting light was frequency-doubled to 420 nm with a β-barium borate crystal to excite the Soret band of chlorophyll a. One portion of the light was directed to a photodiode to become the SYNC pulse for the time-correlated single-photon counting card (Becker-Hickl SPC-630 and SPC-850). The other portion of the laser light was intermittently blocked by a shutter. The average power of the laser at the sample was 5 mW with a pulse energy of 66 pJ. Detection of fluorescence is centered at 684 nm. In addition to the laser light, actinic light (Schott KL1500) with an intensity of 500 or 1,200 μmol photons⋅m−2⋅s−1 as indicated was used to illuminate the sample. The exposure of the leaf to the actinic light was also controlled by a shutter. The dark-adapted leaves were exposed to the actinic light for 45 min. After this first illumination, the actinic light was turned off for 3 min. Then, the light was turned on for a second time for 10 min. The 65 fluorescence lifetime measurements were made at intervals ranging from 3 to 60 s, depending on how fast the fluorescence lifetime was changing. The laser exposure time was 1 s per measurement, divided up into five steps of 0.2 s. To ensure that PSII reaction centers were closed, only the data from the step with the longest fluorescence lifetime were used. This will be discussed further in a future publication.
Twenty leaves from six to eight plants were used for each genotype. The resulting data were aligned according to the maximum of a cross-correlation [xcorAlign.m (27)], and then summed. The fluorescence decay curves were fit to a sum of exponentials (Picoquant Fluofit Pro-4.5). Curves were each fit to three decay functions and one fast rise (1–4 ps), as described by Eq. 1. This fast rise was below the instrument response function of the detector, but was needed to appropriately fit the decays. Following data fitting, curves were reconstructed by plotting the fluorescence intensity calculated by the exponential decays:
| [1] |
where are the amplitudes and are the fluorescence lifetime components. Amplitude-weighted average lifetimes of the leaves were calculated as follows:
| [2] |
The SD of the values of were calculated by performing bootstrapping on the lifetimes. The amplitudes were bootstrapped separately to determine the error in the amplitudes of each component.
Pigment Analysis.
HPLC analysis of xanthophyll content was done as previously described (28). A total of two samples from a set of plants grown at the same time were measured. Leaves were detached from several individual plants of the same genotype and exposed on moist filter paper to a light intensity of 500 μmol photons⋅m−2⋅s−1 for the indicated times. Xanthophylls were quantified using standard curves of purified pigments and normalized to chlorophyll a.
Supplementary Material
Acknowledgments
This material is based upon work supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, Chemical Sciences, Geosciences, and Biosciences Division. E.J.S.-G. was partially supported by a National Science Foundation Graduate Research Fellowship Program. K.K.N. is an investigator of the Howard Hughes Medical Institute and the Gordon and Betty Moore Foundation (through Grant GBMF3070).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1418317111/-/DCSupplemental.
References
- 1.Külheim C, Agren J, Jansson S. Rapid regulation of light harvesting and plant fitness in the field. Science. 2002;297(5578):91–93. doi: 10.1126/science.1072359. [DOI] [PubMed] [Google Scholar]
- 2.Li X-P, Muller-Moule P, Gilmore AM, Niyogi KK. PsbS-dependent enhancement of feedback de-excitation protects photosystem II from photoinhibition. Proc Natl Acad Sci USA. 2002;99(23):15222–15227. doi: 10.1073/pnas.232447699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Avenson TJ, Cruz JA, Kramer DM. Modulation of energy-dependent quenching of excitons in antennae of higher plants. Proc Natl Acad Sci USA. 2004;101(15):5530–5535. doi: 10.1073/pnas.0401269101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wraight CA, Crofts AR. Energy-dependent quenching of chlorophyll alpha fluorescence in isolated chloroplasts. Eur J Biochem. 1970;17(2):319–327. doi: 10.1111/j.1432-1033.1970.tb01169.x. [DOI] [PubMed] [Google Scholar]
- 5.Pfundel EE, Dilley RA. The pH dependence of violaxanthin deepoxidation in isolated pea chloroplasts. Plant Physiol. 1993;101(1):65–71. doi: 10.1104/pp.101.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li X-P, et al. A pigment-binding protein essential for regulation of photosynthetic light harvesting. Nature. 2000;403(6768):391–395. doi: 10.1038/35000131. [DOI] [PubMed] [Google Scholar]
- 7.Niyogi KK, Grossman AR, Björkman O. Arabidopsis mutants define a central role for the xanthophyll cycle in the regulation of photosynthetic energy conversion. Plant Cell. 1998;10(7):1121–1134. doi: 10.1105/tpc.10.7.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holt NE, et al. Carotenoid cation formation and the regulation of photosynthetic light harvesting. Science. 2005;307(5708):433–436. doi: 10.1126/science.1105833. [DOI] [PubMed] [Google Scholar]
- 9.Li X-P, et al. Regulation of photosynthetic light harvesting involves intrathylakoid lumen pH sensing by the PsbS protein. J Biol Chem. 2004;279(22):22866–22874. doi: 10.1074/jbc.M402461200. [DOI] [PubMed] [Google Scholar]
- 10.Dominici P, et al. Biochemical properties of the PsbS subunit of photosystem II either purified from chloroplast or recombinant. J Biol Chem. 2002;277(25):22750–22758. doi: 10.1074/jbc.M200604200. [DOI] [PubMed] [Google Scholar]
- 11.Horton P, Ruban AV, Wentworth M. Allosteric regulation of the light-harvesting system of photosystem II. Philos Trans R Soc Lond B Biol Sci. 2000;355(1402):1361–1370. doi: 10.1098/rstb.2000.0698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Betterle N, et al. Light-induced dissociation of an antenna hetero-oligomer is needed for non-photochemical quenching induction. J Biol Chem. 2009;284(22):15255–15266. doi: 10.1074/jbc.M808625200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goral TK, et al. Light-harvesting antenna composition controls the macrostructure and dynamics of thylakoid membranes in Arabidopsis. Plant J. 2012;69(2):289–301. doi: 10.1111/j.1365-313X.2011.04790.x. [DOI] [PubMed] [Google Scholar]
- 14.Johnson MP, Ruban AV. Restoration of rapidly reversible photoprotective energy dissipation in the absence of PsbS protein by enhanced ΔpH. J Biol Chem. 2011;286(22):19973–19981. doi: 10.1074/jbc.M111.237255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson MP, Zia A, Ruban AV. Elevated ΔpH restores rapidly reversible photoprotective energy dissipation in Arabidopsis chloroplasts deficient in lutein and xanthophyll cycle activity. Planta. 2012;235(1):193–204. doi: 10.1007/s00425-011-1502-0. [DOI] [PubMed] [Google Scholar]
- 16.Lakowicz JR. 2010. Principles of Fluorescence Spectroscopy (Springer, New York), 3rd Ed.
- 17.Nishio JN, Whitmarsh J. Dissipation of the proton electrochemical potential in intact chloroplasts (II. The pH gradient monitored by cytochrome f reduction kinetics) Plant Physiol. 1993;101(1):89–96. doi: 10.1104/pp.101.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nilkens M, et al. Identification of a slowly inducible zeaxanthin-dependent component of non-photochemical quenching of chlorophyll fluorescence generated under steady-state conditions in Arabidopsis. Biochim Biophys Acta. 2010;1797(4):466–475. doi: 10.1016/j.bbabio.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 19.Demmig-Adams B, Adams WW., III Xanthophyll cycle and light stress in nature: Uniform response to excess direct sunlight among higher plant species. Planta. 1996;198(3):460–470. [Google Scholar]
- 20.Havaux M. Carotenoids as membrane stabilizers in chloroplasts. Trends Plant Sci. 1998;3:147–151. [Google Scholar]
- 21.Johnson MP, Ruban AV. Rethinking the existence of a steady-state Δψ component of the proton motive force across plant thylakoid membranes. Photosynth Res. 2014;119(1-2):233–242. doi: 10.1007/s11120-013-9817-2. [DOI] [PubMed] [Google Scholar]
- 22.Cruz JA, Sacksteder CA, Kanazawa A, Kramer DM. Contribution of electric field (Δψ) to steady-state transthylakoid proton motive force (pmf) in vitro and in vivo. Control of pmf parsing into Δψ and ΔpH by ionic strength. Biochemistry. 2001;40(5):1226–1237. doi: 10.1021/bi0018741. [DOI] [PubMed] [Google Scholar]
- 23.Takizawa K, Cruz JA, Kanazawa A, Kramer DM. The thylakoid proton motive force in vivo. Quantitative, non-invasive probes, energetics, and regulatory consequences of light-induced pmf. Biochim Biophys Acta. 2007;1767(10):1233–1244. doi: 10.1016/j.bbabio.2007.07.006. [DOI] [PubMed] [Google Scholar]
- 24.Kramer DM, Sacksteder CA. A diffused-optics flash kinetic spectrophotometer (DOFS) for measurements of absorbance changes in intact plants in the steady-state. Photosynth Res. 1998;56(1):103–112. doi: 10.1023/A:1017906626288. [DOI] [PubMed] [Google Scholar]
- 25.Havaux M, Niyogi KK. The violaxanthin cycle protects plants from photooxidative damage by more than one mechanism. Proc Natl Acad Sci USA. 1999;96(15):8762–8767. doi: 10.1073/pnas.96.15.8762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amarnath K, Zaks J, Park SD, Nigoyi KK, Fleming GR. Fluorescence lifetime snapshots reveal two rapidly reversible mechanisms of photoprotection in live cells of Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 2012;109(22):8405–8410. doi: 10.1073/pnas.1205303109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carmichael J. 2010 xcorAlign.m. MATLAB Central File Exchange. Available at www.mathworks.com/matlabcentral/fileexchange/27456-xcoralign-m/content/xcorAlign.m. Accessed July 16, 2012.
- 28.Müller-Moulé P, Conklin PL, Niyogi KK. Ascorbate deficiency can limit violaxanthin de-epoxidase activity in vivo. Plant Physiol. 2002;128(3):970–977. doi: 10.1104/pp.010924. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.