Abstract
The purpose of this study was to examine the occurrence of fosfomycin-resistant Escherichia coli from chickens and to characterize the plasmids carrying fosA3. A total of 661 E. coli isolates of chicken origin collected from 2009 to 2011 were screened for plasmid-mediated fosfomycin resistance determinants by PCR. Plasmids were characterized using PCR-based replicon typing, plasmid multilocus sequence typing, and restriction fragment length polymorphisms. Associated addiction systems and resistance genes were identified by PCR. PCR-mapping was used for analysis of the genetic context of fosA3. Fosfomycin resistance was detected in 58 isolates that also carried the fosA3 gene. Fifty-seven, 17, and 52 FosA3-producers also harbored blaCTX−M, rmtB, and floR genes, respectively. Most of the 58 fosA3-carrying isolates were clonally unrelated, and all fosA3 genes were located on plasmids belonged to F33:A-:B- (n = 18), IncN-F33:A-:B- (n = 7), IncHI2/ST3 (n = 10), IncI1/ST71 (n = 3), IncI1/ST108 (n = 3), and others. The genetic structures, IS26-ISEcp1-blaCTX−M−55-orf477-blaTEM-1-IS26-fosA3-1758bp-IS26 and ISEcp1-blaCTX−M−65-IS903-iroN-IS26-fosA3-536bp-IS26 were located on highly similar F33:A-:B- plasmids. In addition, blaCTX−M−14-fosA3-IS26 was frequently present on similar IncHI2/ST3 plasmids. IncFII plasmids had a significantly higher frequency of addiction systems (mean 3.5) than other plasmids. Our results showed a surprisingly high prevalence of fosA3 gene in E. coli isolates recovered from chicken in China. The spread of fosA3 can be attributed to horizontal dissemination of several epidemic plasmids, especially F33:A-:B- plasmids. Since coselection by other antimicrobials is the major driving force for the diffusion of the fosA3 gene, a strict antibiotic use policy is urgently needed in China.
Keywords: Escherichia coli, poultry, fosfomycin, plasmids, animal reservoirs, CTX-M
Introduction
The increasing occurrence of bacterial infections caused by multidrug resistant gram-negative Enterobacteriaceae (MDR-GNB) has reignited interest in the old antibiotic, fosfomycin (Falagas et al., 2010). In recent years, fosfomycin has been widely recommended for treating uncomplicated urinary tract infection (UTI) due to its ease of administration and powerful bactericidal activity against MDR-GNB, especially extended-spectrum β-lactamase (ESBL)-producing and fluoroquinolone-resistant Escherichia coli (Falagas et al., 2010; Gardiner et al., 2014; Karlowsky et al., 2014). At the same time, occasional reports of fosfomycin-resistant E. coli strains have emerged (Lee et al., 2012; Ho et al., 2013a; Lai et al., 2014). Mutations in chromosomal genes are the main mechanism for fosfomycin resistance in E. coli (Michalopoulos et al., 2011). However, the transferable fosfomycin resistance genes, fosA, fosC2, and fosK, were recently identified in Asian countries (Wachino et al., 2010; Hou et al., 2012, 2013; Lee et al., 2012; Ho et al., 2013a,b; Kitanaka et al., 2014). Although fosfomycin use in animals is prohibited in China, a high prevalence of the plasmid-mediated fosfomycin-resistance gene fosA3 was observed in E. coli isolates from pet in China (Hou et al., 2012). We also detected fosA3 in E. coli isolates from food animal recovered from 2004 to 2008, but with a relatively lower prevalence compared with pet isolates (Hou et al., 2013). fosA3 is always co-transferred with blaCTX−M genes and the dissemination of fosA3 among pet isolates may be attributed to co-selection by cephalosporins (Hou et al., 2012, 2013). The frequency of blaCTX−M in E. coli isolated from food animals in China has generally been reported to be low prior to 2008, but has increased in prevalence in recent years, especially in chicken isolates (Liu et al., 2007; Li et al., 2010; Zheng et al., 2012). To determine if the occurrence of fosfomycin resistance has also increased with the increasing frequency of blaCTX−M in food animal isolates in recent years, we screened E. coli isolates of chicken origin collected during 2009–2011 for fosfomycin resistance and plasmid-mediated fosfomycin resistance genes. Characterization of fosA3-encoding plasmids as well as the association of fosA3 with other resistance genes, such as blaCTX−M, was also examined.
Materials and methods
Bacterial isolates
A total of 661 E. coli isolates were collected from healthy or diseased chickens in China from 2009 to 2011. Two hundred and ten E. coli from sick chickens at 57 farms were recovered from clinical samples submitted to diagnostic laboratories in Guangdong, Anhui, and Shandong province. The remaining 451 isolates were obtained from fecal samples of healthy chickens from 33 chicken farms located in different geographic regions of China, including Jiangsu, Henan, Guangxi, Gansu, and Guangdong provinces. Sample collection, culture, and E. coli isolation were performed as described previously (Liu et al., 2007; Zheng et al., 2012). E. coli isolates were identified by standard biochemical tests. Assumed fosfomycin- resistant E. coli isolates were selected using Mueller–Hinton (MH) agar plates supplemented with 128 mg/L fosfomycin (Northeast Pharmaceutical Group, Ltd) and 25 μg/mL glucose-6-phosphate and subjected to further study.
Antimicrobial susceptibility testing
Antimicrobial susceptibility test was performed by the agar dilution method on Mueller–Hinton agar plates. The antimicrobial drugs tested included cefotaxime, fosfomycin, gentamicin, amikacin, florfenicol, tetracycline, ciprofloxacin, colistin, and imipenem. Moreover, the isolates were investigated for resistance to tigecycline (15 μg), and piperacillin/tazobactam (110 μg) by the agar disk diffusion method. Both susceptibility tests were carried out and evaluated according to the protocols recommended in VET01-S2 and M100-S23 of the Clinical and Laboratory Standards Institute (2013a,b). E. coli ATCC 25922 was used as the control strain.
Detection of antimicrobial resistance genes
The assumed fosfomycin resistant E. coli isolates were screened for the presence of the plasmid-mediated fosfomycin resistance genes fosA3, fosC2, and fosA by PCR amplification using primers described previously (Ho et al., 2013b). PCR results were confirmed by sequencing. The fosA3-positive strains were also evaluated for the presence of blaCTX−M, rmtB, and floR genes by PCR (Chen et al., 2004, 2007; Sun et al., 2010). The genotype of blaCTX−M was confirmed by PCR and DNA sequencing.
Strain typing
Pulse-field gel electrophoresis (PFGE) of XbaI digested genomic DNA was carried out as described previously (Gautom, 1997) using the CHEF-MAPPER System (Bio-Rad Laboratories, CA, USA). PFGE patterns were analyzed with BioMumerics software (Applied Maths) and were interpreted according to the well-established criteria described by Tenover (Tenover et al., 1995). Isolates that had PFGE patterns with no more than six different bands were considered clonally related. Isolates were further analyzed by multilocus sequence typing (MLST), which was performed according to the specifications given at http://mlst.warwick.ac.uk. MLST profiles were analyzed by Bionumerics.
Conjugation experiments and plasmid analysis
The transferability of fosA3 genes was investigated by conjugation experiments using streptomycin-resistant E. coli C600 as the recipient strain. Transconjugants were selected on MacConkey agar plates supplemented with fosfomycin (200 μg/mL) and streptomycin (2000 μg/mL). Transfer frequencies were calculated as the number of transconjugants per recipient. Transformation experiments were performed to obtain a single plasmid carrying fosA3, as verified by S1 nuclease PFGE when more than one plasmid was co-transferred. The antimicrobial susceptibility of the transconjugants/transformants was determined by the agar dilution method, and the presence of fosA3, blaCTX−M, rmtB, and floR in the transconjugants/transformants was confirmed by PCR. PCR-based replicon typing (PBRT) was performed on all transconjugants/transformants carrying a single plasmid, as described by Carattoli et al. (2005). To better characterize IncFII, IncI, and IncHI2 plasmids, replicon sequence typing (RST), plasmid multilocus sequence typing (pMLST), and plasmid double locus sequence typing (pDLST) were performed according to the procedure described previously (Garcia-Fernandez et al., 2008; Garcia-Fernandez and Carattoli, 2010; Villa et al., 2010), and alleles were assigned by submitting the amplicon sequence to the plasmid MLST database (www.pubmlst.org/plasmid/). F33:A-:B-, IncI1, and IncHI2 plasmids carrying fosA3 were extracted by a rapid alkaline lysis procedure (Takahashi and Nagano, 1984) and further compared using restriction fragment length polymorphism (RFLP) analysis with EcoRI restriction enzymes.
Analysis of the genetic environment of the fosA3 gene
The genetic context surrounding the fosA3 gene was investigated by PCR mapping and sequencing. The primers used to determine the regions upstream and downstream of the fosA3 genes are listed in Table S1.
Nucleotide sequence accession numbers
The new surrounding region of the fosA3 gene found in this study has been deposited in the GenBank database under the following accession numbers: KJ668701 and KJ668702.
Results
Prevalence of plasmid-mediated fosfomycin resistance genes
Of the 661 E. coli isolates from chicken examined, 58 (8.8%) showed resistance to fosfomycin and carried the fosA3 gene. These 58 isolates were recovered from chickens in 37 different farms located in five provinces (Guangdong, Anhui, Shandong, Guangxi, and Henan).
Characterization of fosA3-carrying E. coli isolates
PFGE was successfully performed on 56 E. coli isolates carrying fosA3, and 52 different XbaI PFGE patterns were identified (Table 1). Forty-four different sequence types (STs) were detected among 58 fosA3-positive isolates, including 16 new STs (Table 1 and Supplementary Figure S1). Isolates belonging to ST48 (ST10 clonal complex) were detected in 6 isolates from 6 farms. Minimum spanning tree of MLST of 58 fosA3-carrying E. coli isolates by Bionumerics.
Table 1.
Characterization of fosA3-carrying E. coli isolates.
| Isolatea | Farmb | Isolation date | PFGE patternc | MLST | Resistance profiled |
|---|---|---|---|---|---|
| AHC8 | F1 | 2011.6 | 1 | ST4472 | AMK, CTX, GEN, CIP, FFC, TET |
| AHC9 | F1 | 2011.6 | 2 | ST48 | CTX, GEN, CIP, TET |
| AHC12 | F1 | 2011.6 | 3 | ST533 | CTX, GEN, CIP, FFC, TET |
| AHC16 | F2 | 2011.6 | 3 | ST533 | CTX, GEN, CIP, FFC, TET |
| AHC19 | F4 | 2011.6 | 3 | ST533 | CTX, GEN, CIP, FFC, TET |
| AHC14 | F1 | 2011.6 | 4 | ST746 | CTX, CIP, FFC, TET |
| AHC17 | F2 | 2011.6 | 5 | ST4483 | AMK, CTX, GEN, CIP, FFC, TET |
| AHC18 | F3 | 2011.6 | 6 | ST4447 | CTX, CIP, TET |
| AHC23 | F5 | 2011.6 | 7 | ST2607 | AMK, CTX, GEN, CIP, FFC, TET |
| AHC24 | F5 | 2011.6 | 8 | ST155 | AMK, CTX, GEN, CIP, FFC, TET |
| AHC26 | F6 | 2011.6 | 9 | ST23 | AMK, CTX, GEN, CIP, TET |
| AHC27 | F6 | 2011.6 | 10 | ST2179 | CTX, GEN, CIP, FFC, TET |
| AHC30 | F6 | 2011.6 | 11 | ST226 | CTX, CIP, FFC, TET |
| AHC33 | F7 | 2011.6 | 12 | ST101 | AMK, CTX, GEN, CIP, FFC, TET |
| AHC51 | F9 | 2011.7 | 13 | ST57 | CTX, GEN, CIP, FFC, TET |
| AHC52 | F9 | 2011.7 | 14 | ST206 | AMK, CTX, GEN, CIP, FFC, TET |
| AHC76 | F8 | 2011.7 | 15 | ST4466 | AMK, CTX, GEN, CIP, FFC, TET |
| AHC54 | F9 | 2011.7 | 16 | ST155 | AMK, CTX, GEN, CIP, TET |
| AHC55 | F9 | 2011.7 | 17 | ST162 | CTX, GEN, CIP, FFC, TET |
| AHC57 | F10 | 2011.7 | 18 | ST10 | AMK, CTX, GEN, FFC, TET |
| AHC60 | F11 | 2011.7 | 19 | ST48 | AMK, GEN, CIP, FFC, TET |
| AHC66 | F12 | 2011.7 | 20 | ST48 | CTX, GEN, CIP, FFC, TET |
| AHC67 | F12 | 2011.7 | 21 | ST2223 | CTX, GEN, FFC, TET |
| AHC69 | F13 | 2011.7 | 22 | ST2847 | CTX, CIP, FFC, TET |
| AHC72 | F13 | 2011.7 | 23 | ST2847 | CTX, CIP, FFC, TET |
| AHC80 | F14 | 2011.7 | 24 | ST155 | AMK, CTX, GEN, CIP, FFC, TET |
| GDC27 | F15 | 2010.8 | 25 | ST156 | CTX, GEN, FFC, TET |
| GDC40 | F16 | 2010.8 | 26 | ST744 | CTX, GEN, CIP, FFC, TET |
| GDC54 | F18 | 2010.8 | 27b | ST2496 | CTX, GEN, CIP, FFC, TET |
| GDC46 | F17 | 2010.8 | 27a | ST4460 | CTX, GEN, CIP, FFC, TET |
| GDC47 | F17 | 2010.8 | 28 | ST746 | CTX, CIP, FFC, TET |
| GDC114* | F28 | 2010.9 | 29 | ST48 | AMK, CTX, GEN, CIP, FFC, TET |
| GDC56 | F19 | 2010.8 | 30a | ST359 | CTX, GEN, CIP, FFC, TET |
| GDC58 | F23 | 2010.8 | 30b | ST4461 | CTX, CIP, FFC, TET |
| GDC61 | F20 | 2010.8 | 31 | ST4473 | CTX, GEN, FFC, TET |
| GDC15 | F21 | 2010.8 | 32 | ST4465 | CTX, CIP, FFC |
| GDC16 | F21 | 2010.8 | 33 | ST4474 | AMK, CTX, GEN, CIP, FFC, TET |
| GDC17 | F21 | 2010.8 | 34 | ST1518 | CTX, CIP, FFC, TET |
| GDC24 | F15 | 2010.8 | 35 | ST4477 | CTX, CIP, FFC, TET |
| GDC37 | F22 | 2010.8 | 36 | ST4459 | CTX, GEN, FFC, TET |
| GXC03* | F24 | 2009.7 | 37 | ST2847 | CTX, GEN, CIP, FFC, TET |
| GXC19* | F25 | 2009.7 | 38 | ST4360 | CTX, GEN, CIP, FFC, TET |
| HNC02* | F31 | 2009.7 | 39 | ST4464 | AMK, CTX, GEN, FFC, TET |
| HNC06* | F31 | 2009.7 | 40 | ST779 | CTX, TET |
| SDC20 | F35 | 2009.6 | 41 | ST48 | CTX, FFC, TET |
| SDC10 | F32 | 2009.6 | 42 | ST4498 | AMK, CTX, GEN, CIP, FFC, TET |
| SDC12 | F32 | 2009.6 | 43 | ST4497 | AMK, CTX, GEN, CIP, FFC, TET |
| SDC04 | F33 | 2009.5 | 44 | ST602 | CTX, CIP, FFC, TET |
| SDC01 | F34 | 2009.5 | 45 | ST4462 | AMK, CTX, GEN, CIP, FFC, TET |
| SDC11 | F34 | 2009.6 | 46 | ST602 | AMK, CTX, GEN, CIP, TET |
| SDC15 | F36 | 2009.5 | 47 | ST219 | AMK, CTX, GEN, CIP, FFC, TET |
| GD326 | F27 | 2011.3 | 48 | ST1589 | CTX, CIP, FFC, TET |
| GDC1-4* | F28 | 2010.9 | 49 | ST453 | CTX, GEN, CIP, FFC, TET |
| GDC240* | F29 | 2010.8 | 50 | ST354 | AMK, CTX, GEN, CIP, FFC, TET |
| GDC540* | F29 | 2010.8 | 51 | ST48 | CTX, GEN, FFC, TET |
| GDC1-2* | F28 | 2010.9 | 52 | ST93 | CTX, GEN, CIP, FFC, TET |
| GDC127* | F30 | 2010.9 | smeared | ST156 | CTX, GEN, CIP, FFC, TET |
| SDC13 | F37 | 2009.5 | smeared | ST398 | CTX, GEN, CIP, FFC, TET |
Different provinces are indicated as follows: AH, Anhui; GD, Guangdong; GX, Guangxi; HN, Henan; SD, Shandong. Isolates from which the fosA3 gene can be transferred to the recipient by conjugation are underlined. Healthy animals are indicated by an asterisk.
F1 to F36, farm 1 to farm 36, respectively.
PFGE types (1, 2, 3, etc.) were assigned by visual inspection of the macrorestriction profile. Patterns that differed by fewer than six bands were considered to represent subtypes within the main group (30a, 30b, etc.). NT, nontypeable.
AMK, amikacin; CTX, cefotaxime; CIP, ciprofloxacin; FFC, florfenicol; GEN, gentamicin; TET, tetracycline. The antimicrobial susceptibility results were interpreted according to breakpoint of CLSI (M100-S23), except that florfenicol (≥32 μg/mL) was interpreted according to breakpoint of European Committee on Antimicrobial Susceptibility Testing (EUCAST). All isolates were susceptible to colistin, imipenem, piperacillin-tazobactam, and tigecycline. Resistance phenotypes transferred to the recipient by conjugation ortransformationare underlined.
Of the 58 FosA3-producing E. coli isolates examined in this study, 58 (100%), 57 (98.3%), 57 (98.3%), 52 (89.7%), 49 (84.5%), 44 (75.9%), and 21 (36.2%) were resistant to ampicillin, cefotaxime, tetracycline, florfenicol, ciprofloxacin, gentamicin, and amikacin, respectively (Table 1). All isolates were susceptible to colistin, imipenem, piperacillin-tazobactam, and tigecycline.
Results of screening for resistance genes showed that 57 of the 58 FosA3-producing E. coli isolates carried blaCTX−M genes, including blaCTX−M−55 (n = 24), blaCTX−M−65 (n = 20), blaCTX−M−14 (n = 11), blaCTX−M−123 (n = 3), blaCTX−M−3 (n = 2), blaCTX−M−64 (n = 1), and blaCTX−M−15(n = 1). Five isolates carried two different blaCTX−M genes. In addition, 17 and 52 isolates harbored rmtB and floR genes, respectively.
Analysis of fosA3 plasmids
fosA3 genes from 50 isolates were successfully transferred by conjugation. The fosA3-bearing plasmids in the remaining eight isolates and six transconjugants carrying multiple plasmids were transferred by transformation. Three transformants carried multiple plasmids and were not studied further. The 55 fosA3 plasmids ranged in size from 45 to 230 kb and contained IncFII (n = 29), IncI1 (n = 9), IncHI2 (n = 12), and IncN (n = 4) replicons (Table 2). In addition, seven plasmids were fused plasmids, which contained both IncN and IncFII replicons. The replicon type for one plasmid could not be determined by the PBRT method. Interestingly, the majority of IncFII plasmids associated with the fosA3 gene were classified as F33:A-:B- by RST. Subtyping of IncI1 plasmids revealed three sequence types, including ST71 (n = 3), ST108 (n = 3), and a new sequence type ST136 (n = 2). The IncI1 plasmid from isolate AHC60 was not typable because the trbA and pilL alleles were not detected. Ten IncHI2 plasmids were assigned to ST3 by pDLST, while the other two IncHI2 plasmids were not typable due to failure to detect the smr0199 loci. By restriction analysis of plasmid DNA using EcoRI, 21 F33:A-:B- plasmids, eight IncI1 plasmids, and nine IncHI2 plasmids were divided into 10, eight, and nine groups, respectively, which exhibited small band differences. The conjugation frequencies of F33:A-:B- and IncN-F33:A-:B-plasmids were 10−6 to 10−8, while IncI1 and IncHI2 plasmids were 10−5 to 10−6.
Table 2.
Characterization of some plasmids carrying fosA3.
| Plasmid(s) | Co-transfer of other resistance gene(s) | Context of fosA3a | Plasmid | |||
|---|---|---|---|---|---|---|
| Size (kb) | Replicon type | EcoRI RFLP b | Addiction systems | |||
| GDC24, GDC58, AHC18, SDC13, GDC40, GDC47, GDC1-4, GDC114, SDC04 | blaCTX−M−55 | I | ~75 | F33:A-:B- | A1 | hok-sok, pemKI, srnBC |
| AHC33, GDC240 | blaCTX−M−65, rmtB | III | ~75 | F33:A-:B- | A1 | hok-sok, pemKI, srnBC |
| AHC17, AHC26 | blaCTX−M−55, rmtB | I | ~100 | N-F33:A-:B- | A2 | hok-sok, pemKI, srnBC, vagCD |
| AHC24 | blaCTX−M−55, rmtB, floR | I | ~110 | N-F33:A-:B- | A3 | hok-sok, pemKI, srnBC, vagCD |
| GDC54 | blaCTX−M−65 | III | ~75 | F33:A-:B- | A4 | hok-sok, pemKI, srnBC, vagCD |
| AHC23 | blaCTX−M−65, rmtB | III | ~80 | F33:A-:B- | A5 | hok-sok, pemKI, srnBC |
| GDC46 | blaCTX−M−55 | III | ~75 | F33:A-:B- | A6 | hok-sok, pemKI, srnBC, vagCD |
| SDC01 | blaCTX−M−55, rmtB | I | ~100 | N-F33:A-:B- | A7 | hok-sok, pemKI, srnBC, vagCD |
| AHC76 | blaCTX−M−55 | I | ~75 | F33:A-:B- | A8 | hok-sok, pemKI, srnBC |
| HNC02 | blaCTX−M−65, rmtB | III | ~80 | F33:A-:B- | A9 | hok-sok, pemKI, srnBC, vagCD |
| AHC52 | blaCTX−M−55, rmtB, floR | I | ~110 | N-F33:A-:B- | A10 | hok-sok, pemKI, srnBC, vagCD |
| GDC17 | blaCTX−M−55, floR | I | ~95 | N-F33:A-:B- | B | hok-sok, pemKI, srnBC, vagCD |
| AHC9 | blaCTX−M−55 | I | ~65 | N-F33:A-:B- | C | pemKI, srnBC, vagCD |
| AHC69, AHC72 | blaCTX−M−65 | I | ~75 | F33:A-:B- | ND | hok-sok, pemKI, srnBC, vagCD |
| AHC27 | blaCTX−M−65 | I | ~115 | I1/ST71 | D1 | pemKI, pndCA, vagCD |
| GDC27 | blaCTX−M−65, floR | I | ~125 | I1/ST71 | D2 | pemKI |
| AHC30 | blaCTX−M−55, floR | I | ~105 | I1/ST136 | D3 | hok-sok, pemKI, pndCA, vagCD |
| GXC19 | blaCTX−M−65, floR | II | ~125 | I1/ST71 | D4 | pndCA |
| AHC54 | blaCTX−M−123 | I | ~110 | I1/ST108 | D5 | pemKI, pndCA, vagCD |
| AHC14 | blaCTX−M−123 | I | ~115 | I1/ST108 | D6 | pndCA |
| AHC55 | blaCTX−M−123 | I | ~110 | I1/ST108 | D7 | hok-sok, pemKI, pndCA |
| SDC11 | blaCTX−M−14, rmtB | V | ~100 | I1/ST136 | D8 | pemKI, pndCA |
| AHC60 | rmtB | Unknown | ~120 | I1 | E | pndCA, vagCD |
| AHC66, AHC67 | blaCTX−M−65, floR | I | ~230 | HI2/ST3 | F1 | none |
| AHC57 | blaCTX−M−14, floR | IV | ~230 | HI2/ST3 | F2 | none |
| AHC80 | blaCTX−M−14, rmtB, floR | IV | ~230 | HI2/ST3 | ND | vagCD |
| GDC540 | blaCTX−M−14, floR | V | ~230 | HI2/ST3 | F3 | pemKI, vagCD |
| GDC15 | blaCTX−M−65, floR | III | ~230 | HI2/ST3 | F4 | none |
| GDC61 | blaCTX−M−14, floR | IV | ~230 | HI2 | F5 | pemKI, vagCD |
| GXC03 | blaCTX−M−15 | Unknown | ~230 | HI2/ST3 | F6 | vagCD |
| GDC127 | blaCTX−M−14, floR | V | ~230 | HI2/ST3 | F7 | none |
| HNC06 | blaCTX−M−14 | V | ~230 | HI2/ST3 | F8 | vagCD |
| GD326 | blaCTX−M−14, floR | V | ~230 | HI2 | F9 | none |
| SDC20 | blaCTX−M−65, floR | I | ~230 | HI2/ST3 | ND | none |
| SDC10, SDC12 | rmtB, floR | I | ~45 | N | ND | none |
| SDC15 | rmtB | I | ~50 | N | ND | vagCD |
| AHC51 | blaCTX−M−65 | I | ~50 | N | ND | none |
| GDC1-2 | blaCTX−M−3, floR | I | ~75 | F2:A-:B- | ND | hok-sok, pemKI, vagCD |
| GDC56 | blaCTX−M−3 | I | ~75 | F14:A-:B- | ND | hok-sok, pemKI, vagCD |
| AHC12 | blaCTX−M−65, floR | I | ~80 | F18:A-:B- | ND | pemKI, srnBC, vagCD, ccdAB |
| AHC16 | blaCTX−M−14, floR | I | ~80 | F18:A-:B- | ND | pemKI, srnBC, ccdAB |
| GDC16 | I | ~70 | unknown | ND | vagCD | |
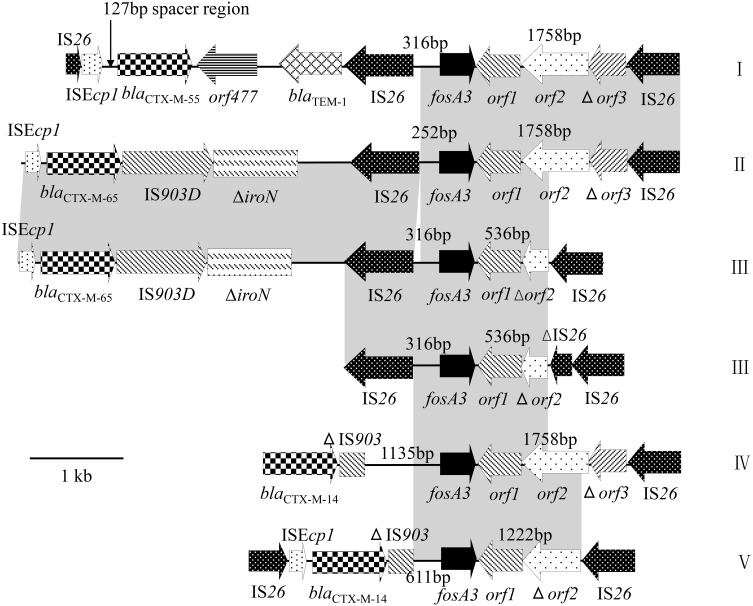
Contexts of fosA3 were as follows: I, IS26-316bp-fosA3-1758bp-IS26; II, IS26-316bp-fosA3-536bp-IS26; III, blaCTX−M−14-1135bp-fosA3-1758bp-IS26; IV, blaCTX−M−14-611bp-fosA3-1222bp-IS26.
Restriction fragment length polymorphism (RFLP) patterns differed by only a few bands (n = 1–3) were assigned to the same RFLP profile. ND, not determined.
Co-transfer of resistance to other antimicrobials (cefotaxime, aminoglycosides, florfenicol, and tetracycline) was observed in 54 of the 55 transconjugants/transformants harboring a single plasmid. blaCTX−M, rmtB, and floR genes were co-transferred with fosA3 to the recipients from 50, 14, and 19 donors, respectively.
Addiction systems of fosA3 plasmids
The 55 fosA3 plasmids carried 0-4 addiction systems (mean 2.4). Six different systems, namely ccdAB, hok-sok, pemKI, pndAC, snrBC, and vagCD systems were detected. The most frequently represented systems were pemKI, followed by vagCD, hok-sok, and srnBC (Table 2). All F33: A-: B- plasmids except one (AHC9) had pemKI, hok-sok, and srnBC. The average number of addiction systems detected was the highest (3.5) among IncFII plasmids, followed by IncI1 plasmids (2.2), which were significantly higher than the remaining plasmids (P < 0.01). IncN and IncHI2 plasmids were mostly devoid of the addiction systems tested in the study (Table 2).
Genetic environment of fosA3
The regions surrounding fosA3 were determined by PCR mapping and sequencing. Different genetic contexts of fosA3 were designated as types I–V (Table 2 and Figure 1). An IS26 element was found to be located downstream of fosA3 in all the isolates except AHC60, AHC8, and GXC03. In these three isolates, the genetic elements downstream of fosA3 could not be defined. The sizes of the spacer regions between the 3′ end of fosA3 and IS26 varied (1758, 536, and 1222 bp; Table 1). Upstream of fosA3, four different genetic organizations were identified. In 48 isolates, IS26 was located 316 bp upstream of fosA3 (type I, III), and in 1 isolate (GXC19), IS26 was located 252 bp upstream of fosA3 (type II). In nine isolates, blaCTX−M−14 was identified 1135 bp upstream of fosA3 (type IV) or 611 bp upstream of fosA3 (type V). The type V structure (blaCTX−M−14-611bp-fosA3-1222bp-IS26) was 100% identical to that found in the plasmids of ECO021TF (accession no. JQ343849, human E. coli, Korea, 2009), pHP48 (AB778503, human E. coli, Japan, 2010), and pN0863T (JQ823170, dog E. coli, Hong Kong, 2008). The type IV structure (blaCTX−M−14-1135bp-fosA3-1758bp-IS26) was 100% identical to that found in the plasmid of EC0121TF (accession no. JX442753, chicken E. coli, 2009) from Hong Kong and was found to be located on IncHI2 plasmids. IncHI2 was also found to be associated with type V structures. However, type I and type III structures were usually found to be associated with F33:A-:B- and IncI1 plasmids. In 15 F33:A-:B- or N-F33:A-:B- plasmids, a structure comprising IS26, truncated ISEcp1, blaCTX−M−55, orf477, and a truncated blaTEM−1, was found upstream of the type I structure. This genetic environment surrounding the fosA3 gene was 100% in sequence identity to the plasmid carried by E. coli HP558 (AB778291, human E. coli, 2010) from Japan. Also, in five F33:A-:B- plasmids and four IncI1 plasmids, a structure comprising IS26, truncated ISEcp1, blaCTX−M−65, IS903, and iroN was found upstream of the type I structure and type III structure, respectively. The genetic structure (ISEcp1-blaCTX−M−65-IS903-iroN-IS26-316bp-fosA3-536bp-IS26) located on F33:A-:B- plasmids was almost identical to those found in plasmids pXZ (JF927996, duck E. coli, 2008) and pHN7A8 (JN232517, dog E. coli, 2008) from China. The genetic structure (ISEcp1-blaCTX−M−65-IS903-iroN-IS26-252bp-fosA3-1758bp-IS26) carried by IncI1 plasmids represented a novel genetic environment and was first identified in this study. In one isolate with the type III fosA3 context, the IS26 downstream of fosA3 was truncated by another IS26.
Figure 1.
Schematic representation of the genetic contexts of fosA3 gene. Regions of 100% homology are shaded in gray.
Discussion
In this study, we investigated the prevalence of fosfomycin resistance in E. coli isolated from chickens from 2009 to 2011 in China. Our previous study showed that fosfomycin resistance was unusual among E. coli isolates from food animals during the period from 2004 to 2008 (Hou et al., 2013). However, in this study, fosfomycin resistance was detected in 8.8% of E. coli isolates from chickens. This frequency was significantly higher than that reported in other countries (Karageorgopoulos et al., 2012; Karlowsky et al., 2014), and also higher than that in isolates from humans in China (Lai et al., 2014). Fosfomycin is not approved for use in food animals in China. Thus, the high frequency of fosfomycin resistance found in this study was unexpected and was not due to the selective pressure resulting from exposure to fosfomycin. Coselection by other antimicrobials, especially third-generation cephalosporins, seemed to play a major role in facilitating the spread of this plasmid-mediated fosfomycin-resistance determinant since almost all fosA3 plasmids also carried blaCTX−M genes. In addition, the intensive use of florfenicol and gentamicin might also have favored the dissemination of fosA3 in the chicken population as florfenicol- and gentamicin-resistance genes were usually co-transferred with fosA3. Other studies also observed the cotransfer of blaCTX−M genes with the fosA3 gene and the cotransfer of chloramphenicol resistance (Hou et al., 2012, 2013; Ho et al., 2013a; Sato et al., 2013; Lai et al., 2014).
Most of the fosA3-positive isolates (48/58) found in this study were recovered from diseased chickens. Generally, chicken farmers will use antimicrobial agents to control diseases; if such disease controls fail, they will send the diseased animals to diagnostic laboratories for diagnosis and treatment. Thus, the diseased chickens have most likely been subjected to more serious antimicrobial selective forces than healthy chickens before they are sent to diagnostic laboratories, suggesting that other antimicrobials may be the driving force for the observed increase in fosfomycin resistance and may affect the dissemination of fosA3 in chickens. In support of this, we surveyed the antimicrobial usage history of 15 farms (data not shown) and found that cephalosporins and florfenicol were frequently used in most chicken farms.
The spread of the fosA3 gene among E. coli isolates of chicken origin was not attributed to clonal transfer of FosA3-producers, but was instead caused by several epidemic plasmids, including F33:A-:B-, IncI1/ST108, IncI1/ST71, and IncHI2/ST3 plasmids, which have been disseminated in multiple chicken farms found in different geographic regions of China.
The combination of fosA3 and F33:A-:B- plasmids has been frequently identified in several Asia countries (Hou et al., 2012, 2013; Lee et al., 2012; Sato et al., 2013; Pan et al., 2014). In our previous study on isolates from pets and food animals collected during the period from 2004 to 2008, fosA3 genes usually co-existed with baCTX−M−65 on F33:A-:B- plasmids (Hou et al., 2012, 2013). However, in this study, fosA3 genes frequently co-existed with blaCTX−M−55 on a similar or identical genetic structure carried by F33:A-:B- plasmids, similar to that of the two F33:A-:B- plasmids found in Japan and Korea (Lee et al., 2012; Sato et al., 2013). These data indicated that F33:A-:B- plasmids carrying identical or similar mobile multiresistance regions have disseminated in animals and humans in different Asian countries. Therefore, these plasmids are capable of spreading very efficiently and may be the major vehicle contributing to the spread of the fosA3 gene. The successful dissemination of F33:A-:B- plasmids may be attributed to the presence of addiction systems (pemKI, hok-sok, srnBC), which ensure the stable maintenance of the plasmid during cell division (Hou et al., 2012; He et al., 2013b). Though the F33:A-:B- plasmid has only been identified in Asian countries, it may be possible for the plasmid to disseminate worldwide by international travel or animal and food trade.
IncI1 plasmids carrying fosA3 have previously been reported in China and Japan (Hou et al., 2013; Sato et al., 2013). In this study, three different IncI1 pMLST plasmid types were associated with fosA3, namely ST71, ST108, and ST136. Three ST71 plasmids from different provinces (Anhui, Guangxi, and Guangdong) carried both fosA3 and blaCTX−M−65. Interestingly, ST71 carrying blaCTX−M−14 and fosA3 was also detected in isolates from humans in Japan (Sato et al., 2013). ST108, first reported in our previous study as a blaCTX−M−123 carrier (He et al., 2013a), also harbored blaCTX−M−123 in this study. In a pig farm in the United Kingdom, the ST108 IncI1 plasmid carrying the blaCTX−M−1 gene had disseminated across multiple genera (Freire Martin et al., 2014). IncHI2 plasmids have been found to be associated with ESBL genes in Enterobacteriaceae, but were more common in Salmonella enterica than in E. coli (Garcia-Fernandez and Carattoli, 2010). This study reported the identification of fosA3 genes on IncHI2 plasmids for the first time. Most fosA3 genes in IncHI2 plasmids have similar genetic environments, mainly blaCTX−M−14-fosA3-IS26. However, this structure was located on the IncN plasmid in one isolate from Korea (Lee et al., 2012). Our findings revealed that the mobile element IS26 and co-selection with blaCTX−M genes played a critical role in the rapid transfer of the fosA3 gene between diverse epidemic plasmids. Since fosA3 can be carried by several successfully disseminated plasmids (F33:A-:B-, ST108/ST71 IncI1, and ST3 IncHI2) and CTX-M-type ESBL producers are distributed globally in a variety of settings (Woerther et al., 2013), we should pay close attention to the worldwide dissemination of fosA3 in the near future. More studies are required to investigate the spread of fosA3 gene in other countries as well as in other hosts and environments.
In conclusion, our study reported a surprisingly high prevalence of the plasmid-mediated fosfomycin-resistance gene fosA3 in E. coli isolates from chicken in China. Fosfomycin has become one of the limited treatment options for critically ill patients with multidrug-resistant bacteria, especially carbapenem-resistant gram-negative bacteria (Dortet et al., 2014), further spread of the fosA3 gene would be a serious public health concern. Measures must be implemented to avoid the selection and spread of fosfomycin-resistant strains. Since the fosA3 gene is usually cotransferred with blaCTX−M−55, blaCTX−M−65, blaCTX−M−14, floR, and rmtB genes on several epidemic plasmids, reduction in total antimicrobial use, particularly cephalosporins, in food animal production in China may help to control the spread of plasmid-mediated fosfomycin-resistance genes.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported in part by National Key Basic Research Program of China (No. 2013CB127200), the Program for Changjiang Scholars and Innovative Research Team in University (No. IRT13063), the National Natural Science Foundation and Natural Science Foundation of Guangdong Province, China (No.U1031004), and the Foundation for High-level Talents in Higher Education of Guangdong, China.
Supplementary material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fmicb.2014.00688/abstract
References
- Carattoli A., Bertini A., Villa L., Falbo V., Hopkins K. L., Threlfall E. (2005). Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63, 219–228. 10.1016/j.mimet.2005.03.018 [DOI] [PubMed] [Google Scholar]
- Chen L., Chen Z. L., Liu J. H., Zeng Z. L., Ma J. Y., Jiang H. X. (2007). Emergence of RmtB methylase-producing Escherichia coli and Enterobacter cloacae isolates from pigs in China. J. Antimicrob. Chemother. 59, 880–885. 10.1093/jac/dkm065 [DOI] [PubMed] [Google Scholar]
- Chen S., Zhao S., White D. G., Schroeder C. M., Lu R., Yang H., et al. (2004). Characterization of multiple-antimicrobial-resistant salmonella serovars isolated from retail meats. Appl. Environ. Microbiol. 70, 1–7. 10.1128/AEM.70.1.1-7.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical Laboratory Standards Institute. (2013a). Performance Standards for Antimicrobial disk and Dilution Susceptibility Test for Bacteria Isolated from Animals; Second Informational Supplement. CLSI document VET01-S2. Wayne, PA: CLSI. [Google Scholar]
- Clinical Laboratory Standards Institute. (2013b). Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Third Informational Supplement. CLSI document M100-S23. Wayne, PA:CLSI. [Google Scholar]
- Dortet L., Poirel L., Nordmann P. (2014). Worldwide dissemination of the NDM-Type carbapenemases in gram-negative bacteria. BioMed. Res. Int. 2014:249856. 10.1155/2014/249856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falagas M. E., Kastoris A. C., Kapaskelis A. M., Karageorgopoulos D. E. (2010). Fosfomycin for the treatment of multidrug-resistant, including extended-spectrum beta-lactamase producing, Enterobacteriaceae infections: a systematic review. Lancet Infect. Dis. 10, 43–249850. 10.1016/S1473-3099(09)70325-1 [DOI] [PubMed] [Google Scholar]
- Freire Martin I., Abuoun M., Reichel R., La Ragione R. M., Woodward M. J. (2014). Sequence analysis of a CTX-M-1 IncI1 plasmid found in Salmonella 4, 5,12:i:-, Escherichia coli and Klebsiella pneumoniae on a UK pig farm. J. Antimicrob. Chemother. 69, 2098–2101. 10.1093/jac/dku098 [DOI] [PubMed] [Google Scholar]
- Garcia-Fernandez A., Carattoli A. (2010). Plasmid double locus sequence typing for IncHI2 plasmids, a subtyping scheme for the characterization of IncHI2 plasmids carrying extended-spectrum beta-lactamase and quinolone resistance genes. J. Antimicrob. Chemother. 65, 1155–1161. 10.1093/jac/dkq101 [DOI] [PubMed] [Google Scholar]
- Garcia-Fernandez A., Chiaretto G., Bertini A., Villa L., Fortini D., Ricci A., et al. (2008). Multilocus sequence typing of IncI1 plasmids carrying extended-spectrum beta-lactamases in Escherichia coli and Salmonella of human and animal origin. J. Antimicrob. Chemother. 61, 1229–1233. 10.1093/jac/dkn131 [DOI] [PubMed] [Google Scholar]
- Gardiner B. J., Mahony A. A., Ellis A. G., Lawrentschuk N., Bolton D. M., Zeglinski P. T., et al. (2014). Is fosfomycin a potential treatment alternative for multidrug-resistant gram-negative prostatitis? Clin. Infect. Dis. 58, e101–e105. 10.1093/cid/cit704 [DOI] [PubMed] [Google Scholar]
- Gautom R. K. (1997). Rapid pulsed-field gel electrophoresis protocol for typing of Escherichia coli O157:H7 and other gram-negative organisms in 1 day. J. Clin. Microbiol. 35, 2977–2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He D., Partridge S. R., Shen J., Zeng Z., Liu L., Rao L., et al. (2013a). CTX-M-123, a novel hybrid of the CTX-M-1 and CTX-M-9 group beta-Lactamases recovered from Escherichia coli isolates in China. Antimicrob. Agents Chemother. 57, 4068–4071. 10.1128/AAC.00541-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L., Partridge S. R., Yang X., Hou J., Deng Y., Yao Q., et al. (2013b). Complete nucleotide sequence of pHN7A8, an F33:A-:B- type epidemic plasmid carrying blaCTX-M-65, fosA3 and rmtB from China. J. Antimicrob. Chemother. 68, 46–50. 10.1093/jac/dks369 [DOI] [PubMed] [Google Scholar]
- Ho P. L., Chan J., Lo W. U., Lai E. L., Cheung Y. Y., Lau T. C., et al. (2013a). Prevalence and molecular epidemiology of plasmid-mediated fosfomycin resistance genes among blood and urinary Escherichia coli isolates. J. Med. Microbiol. 62, 1707–1713. 10.1099/jmm.0.062653-0 [DOI] [PubMed] [Google Scholar]
- Ho P. L., Chan J., Lo W. U., Law P. Y., Li Z., Lai E. L., et al. (2013b). Dissemination of plasmid-mediated fosfomycin resistance fosA3 among multidrug-resistant Escherichia coli from livestock and other animals. J. Appl. Microbiol. 114, 695–702. 10.1111/jam.12099 [DOI] [PubMed] [Google Scholar]
- Hou J., Huang X., Deng Y., He L., Yang T., Zeng Z., et al. (2012). Dissemination of the fosfomycin resistance gene fosA3 with CTX-M beta-lactamase genes and rmtB carried on IncFII plasmids among Escherichia coli isolates from pets in China. Antimicrob. Agents Chemother. 56, 2135–2138. 10.1128/AAC.05104-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou J., Yang X., Zeng Z., Lv L., Yang T., Lin D., et al. (2013). Detection of the plasmid-encoded fosfomycin resistance gene fosA3 in Escherichia coli of food-animal origin. J. Antimicrob. Chemother. 68, 766–770. 10.1093/jac/dks465 [DOI] [PubMed] [Google Scholar]
- Karageorgopoulos D. E., Wang R., Yu X. H., Falagas M. E. (2012). Fosfomycin: evaluation of the published evidence on the emergence of antimicrobial resistance in Gram-negative pathogens. J. Antimicrob. Chemother. 67, 255–268. 10.1093/jac/dkr466 [DOI] [PubMed] [Google Scholar]
- Karlowsky J. A., Denisuik A. J., Lagace-Wiens P. R., Adam H. J., Baxter M. R., Hoban D. J., et al. (2014). In Vitro activity of fosfomycin against Escherichia coli isolated from patients with urinary tract infections in Canada as part of the CANWARD surveillance study. Antimicrob. Agents Chemother. 58, 1252–1256. 10.1128/AAC.02399-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitanaka H., Wachino J. I., Wanchun J., Yokoyama S., Sasano M. A., Hori M., et al. (2014). Novel integron-mediated fosfomycin-resistance genetic determinant, fosK. Antimicrob. Agents Chemother. 58, 4978–4979. 10.1128/AAC.03131-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai B., Zheng B., Li Y., Zhu S., Tong Z. (2014). In vitro susceptibility of Escherichia coli strains isolated from urine samples obtained in mainland China to fosfomycin trometamol and other antibiotics: a 9-year surveillance study (2004-2012). BMC Infect. Dis. 14:66. 10.1186/1471-2334-14-66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. Y., Park Y. J., Yu J. K., Jung S., Kim Y., Jeong S. H., et al. (2012). Prevalence of acquired fosfomycin resistance among extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae clinical isolates in Korea and IS26-composite transposon surrounding fosA3. J. Antimicrob. Chemother. 67, 2843–2847. 10.1093/jac/dks319 [DOI] [PubMed] [Google Scholar]
- Li J., Ma Y., Hu C., Jin S., Zhang Q., Ding H., et al. (2010). Dissemination of cefotaxime-M-producing Escherichia coli isolates in poultry farms, but not swine farms, in China. Foodborne Pathog. Dis. 7, 1387–1392. 10.1089/fpd.2010.0581 [DOI] [PubMed] [Google Scholar]
- Liu J. H., Wei S. Y., Ma J. Y., Zeng Z. L., Lu D. H., Yang G. X., et al. (2007). Detection and characterisation of CTX-M and CMY-2 beta-lactamases among Escherichia coli isolates from farm animals in Guangdong Province of China. Int. J. Antimicrob. Agents 29, 576–581. 10.1016/j.ijantimicag.2006.12.015 [DOI] [PubMed] [Google Scholar]
- Michalopoulos A. S., Livaditis I. G., Gougoutas V. (2011). The revival of fosfomycin. Int. J. Infect. Dis. 15, e732–e739. 10.1016/j.ijid.2011.07.007 [DOI] [PubMed] [Google Scholar]
- Pan Y. S., Yuan L., Zong Z. Y., Liu J. H., Wang L. F., Hu G. Z. (2014). A multidrug-resistance region containing blaCTX-M-65, fosA3 and rmtB on conjugative IncFII plasmids in Escherichia coli ST117 isolates from chicken. J. Med. Microbiol. 63, 485–488. 10.1099/jmm.0.070664-0 [DOI] [PubMed] [Google Scholar]
- Sato N., Kawamura K., Nakane K., Wachino J., Arakawa Y. (2013). First detection of fosfomycin resistance gene fosA3 in CTX-M-producing Escherichia coli isolates from healthy individuals in Japan. Microb. Drug Resist. 19, 477–482. 10.1089/mdr.2013.0061 [DOI] [PubMed] [Google Scholar]
- Sun Y., Zeng Z., Chen S., Ma J., He L., Liu Y., et al. (2010). High prevalence of bla(CTX-M) extended-spectrum beta-lactamase genes in Escherichia coli isolates from pets and emergence of CTX-M-64 in China. Clin. Microbiol. Infect. 16, 1475–1481. 10.1111/j.1469-0691.2010.03127.x [DOI] [PubMed] [Google Scholar]
- Takahashi S., Nagano Y. (1984). Rapid procedure for isolation of plasmid DNA and application to epidemiological analysis. J. Clin. Microbiol. 20, 608–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenover F. C., Arbeit R. D., Goering R. V., Mickelsen P. A., Murray B. E., Persing D. H., et al. (1995). Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33, 2233–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa L., Garcia-Fernandez A., Fortini D., Carattoli A. (2010). Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J. Antimicrob. Chemother. 65, 2518–2529. 10.1093/jac/dkq347 [DOI] [PubMed] [Google Scholar]
- Wachino J., Yamane K., Suzuki S., Kimura K., Arakawa Y. (2010). Prevalence of fosfomycin resistance among CTX-M-producing Escherichia coli clinical isolates in Japan and identification of novel plasmid-mediated fosfomycin-modifying enzymes. Antimicrob. Agents Chemother. 54, 3061–3064. 10.1128/AAC.01834-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woerther P. L., Burdet C., Chachaty E., Andremont A. (2013). Trends in human fecal carriage of extended-spectrum beta-lactamases in the community: toward the globalization of CTX-M. Clin. Microbiol. Rev. 26, 744–758. 10.1128/CMR.00023-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H., Zeng Z., Chen S., Liu Y., Yao Q., Deng Y., et al. (2012). Prevalence and characterisation of CTX-M beta-lactamases amongst Escherichia coli isolates from healthy food animals in China. Int. J. Antimicrob. Agents 39, 305–310. 10.1016/j.ijantimicag.2011.12.001 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.