Abstract
Objectives
Polymorphisms in the lysosomal transporter encoded by the pfcrt gene directly impact on Plasmodium falciparum susceptibility to aminoquinolines. The Lys76Thr mutation is the critical change conferring chloroquine resistance in vitro and in vivo, but always occurs with additional non-synonymous changes in the pfcrt coding sequence. We sought to better describe pfcrt polymorphisms distal to codon 76.
Methods
Small-volume samples (≤500 μL) of parasite-infected blood collected directly from malaria patients presenting for treatment in Sudan and Tanzania were immediately preserved for RNA extraction. The pfcrt locus was amplified from cDNA preparations by nested PCR and sequenced directly to derive full-length mRNA sequences.
Results
In one of two sites in Sudan, two patients were found with an unorthodox spliced form of pfcrt mRNA in which two exons were skipped, but it was not possible to test for the presence of the putative protein products of these aberrant transcripts. Genomic DNA sequencing from dried blood spots collected in parallel confirmed the presence of spliced pfcrt pseudogenes in a minority of parasite isolates. Full-length cDNA from conventionally spliced mRNA molecules in all study sites demonstrated the existence of a variety of pfcrt haplotypes in East Africa, and thus provides evidence of intragenic recombination.
Conclusions
The presence of pseudogenes, although unlikely to have any direct public health impact, may confound results obtained from simple genotyping methods that consider only codon 76 and the adjacent residues of pfcrt.
Keywords: malaria, chloroquine resistance transporters, alternative splicing
Introduction
Resistance of the malaria parasite Plasmodium falciparum to antimalarial drugs has posed a serious threat to public health in Africa in the past,1 and may do so again in the future.2 Molecular markers of resistance to chloroquine, pyrimethamine and sulfadoxine were identified and validated some years after the efficacy of these drugs had waned; the utility of markers was thus mainly in gaining understanding of events that had already happened. In the context of artemisinin-based combination therapy (ACT) being the mainstay of uncomplicated malaria management worldwide, malaria researchers have recently focused on the identification of candidate markers for diminished efficacy of artemisinin and of the partner drugs used in ACT.3 Such markers are needed as molecular tools for surveillance for the purpose of preventing the global spread of resistant parasites from foci of origin, and this objective has become more urgent since the recent description in Cambodia and neighbouring countries of parasites with reduced susceptibility to artemisinin compounds.4,5
It is now well established that ACT selects for particular alleles of two genes, pfmdr1 and pfcrt, both firmly associated with resistance to the 4-aminoquinolines chloroquine and amodiaquine. In a number of African studies of ACT efficacy in symptomatic children and adults, the aminoquinoline-susceptible haplotypes of these genes were significantly more common in recurrent parasitaemia detected during follow-up after treatment with the most commonly used ACT, artemether/lumefantrine.6–9
Encoding a transporter located in the parasite's digestive vacuole membrane, pfcrt is an important determinant of susceptibility to antimalarial chemotherapy. Variant genes encoding a threonine instead of a lysine at codon 76 (the K76T mutation) are highly resistant to chloroquine in vitro and more likely to be present in parasites surviving chloroquine or amodiaquine treatment in vivo,10–13 but less likely to survive in ACT-treated malaria patients.6–9 The gene encoding the PfCRT protein comprises 13 short exons interrupted by AT-rich introns, imposing technical challenges when sequencing the gene from small-volume, non-replenishable samples. Thus, available sequence data not immediately adjacent to codon 76 have mostly been derived from cDNA obtained from culture-adapted parasite lines.14,15 More information on polymorphisms across the full-length pfcrt gene is therefore needed, particularly from in vivo studies of treated malaria patients. In this study transcripts of the pfcrt gene were isolated from peripheral blood collected directly from symptomatic malaria cases prior to treatment in two East African settings, and characterized by extended cDNA sequencing, in order to reconstruct full genetic haplotypes present in vivo. We report for the first time alternatively spliced forms of pfcrt mRNA, and the presence of genomic intron-free pfcrt ORF sequences, likely to represent processed—and probably transcribed—pseudogenes.
Materials and methods
Study population
Peripheral blood samples for this study were collected as part of a trial of artemether/lumefantrine efficacy in both adults and children, conducted in New Halfa and Gedarif in eastern Sudan in 2006,9 and a pharmacokinetic study of children aged 1–10 years treated with artemether/lumefantrine for uncomplicated P. falciparum infection, performed in Fukayosi, Bagamoyo district, Tanzania in 2006.16,17 Details of the study area, study design, patient recruitment criteria, ethical considerations and treatment outcome have been previously reported for both studies.
Sample collection and nucleic acid preparation
For RNA isolation and cDNA synthesis, in both Sudanese studies whole blood was collected in EDTA tubes prior to treatment. A 60 μL aliquot of each sample was mixed with 500 μL of TRI reagent (Sigma) and stored initially at −20°C and later at −80°C once the field work was completed. RNA was extracted by the TRI reagent protocol or the SV total RNA extraction kit (Promega) as per the manufacturers' instructions and DNA removed by digestion with deoxyribonuclease I. cDNA was synthesized employing the Superscript III kit (Invitrogen) as per the manufacturer's recommendations, using random oligonucleotide hexamers to prime the first-strand synthesis. In Fukayosi, from each patient 0.5 mL of whole blood was collected in ribonuclease-free tubes containing 0.5 mL of PBS and 1 mL of 2× Lysis Solution (Applied Biosystems), mixed by shaking and stored directly on ice, then transported to Karolinska Institutet, Sweden on dry ice and stored at −20°C. RNA isolation was performed using the ABI 6100 machine according to the Applied Biosystems protocol for isolation of total RNA from whole blood. Samples were thereafter stored at −80°C. cDNA was synthesized using random hexamers provided in the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems).
For DNA extraction, pretreatment finger-prick blood samples were collected onto glass fibre membranes (Wallac, Finland), dried and stored at 4°C. DNA was extracted by the Chelex® method as previously described.9
Amplification and sequencing of the pfcrt gene
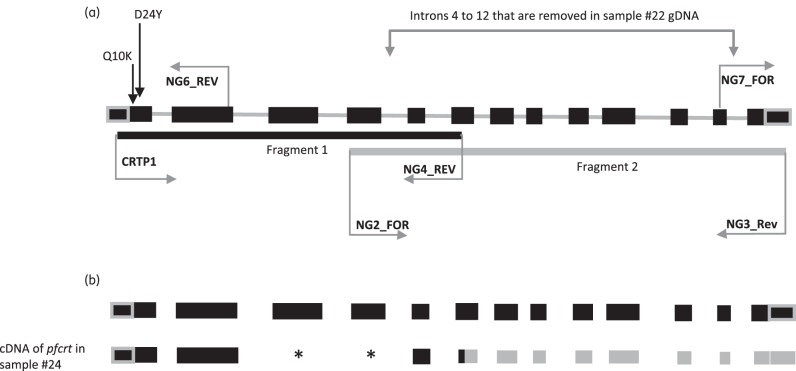
Full-length cDNA and genomic DNA (gDNA) of the pfcrt gene were amplified in two overlapping PCR amplicons. Fragment 1 [86 nt of the 5′ untranslated region (UTR), plus codons 1–247] was amplified with primers CRTP111 and NG1_REV in the outer reaction and semi-nested with NG4_REV for fragment 1. Fragment 2 (codons 220–424, plus 151 nt of the 3′ UTR) was amplified with primers NG5_FOR and NG3_REV for the outer reaction and NG2_FOR and NG3_REV for the nested reaction (Figure 1a). Non-specific and incomplete single-stranded DNA was removed from PCR products using EXO I exonuclease (Fermentas) prior to sequencing on both strands using the amplification primers (ABI BigDye v3 Cycle Terminator kit). In addition, two extra primers (NG6_REV and NG7_FOR; Table 1) were employed at the 5′ and 3′ terminals, respectively, to sequence the full length of the gene. For gDNA, additional sequencing reactions employing primers P11 (on exon), P13 (crosses intron) and P17 (on intron) were performed. Amplification and sequencing primers, and cycling conditions used, are presented in Table 1. Analysis was performed as previously described.9
Figure 1.
Amplification strategy of the pfcrt gene employed in this study. (a) Amplification of the PF3D7_0709000 gDNA of the pfcrt gene, exons (black boxes), introns (solid grey lines) and UTR (grey boxes with black fill). Fragment 1 (solid black line) and fragment 2 (solid grey line) are indicated beneath the gene map. PCR primer pairs used to amplify the whole gene (in both gDNA and cDNA) are indicated by horizontal arrows. Primers NG6_REV and NG7_FOR were used to sequence the UTR. Locations of Q10K and D24Y polymorphisms are indicated with arrows. Intron deletion region in gDNA is indicated. (b) cDNA amplified from the pfcrt gene. Upper transcript map is the reference PF3D7_0709000 pfcrt cDNA. Lower map is the variant pfcrt cDNA amplified in sample 24. Black rectangles represent the sequenced transcribed exons. Asterisks represent the non-transcribed (spliced) exons. Grey rectangles represent the putative mRNA downstream from codon 220; this region was not sequenced from cDNA but inferred from gDNA sequence.
Table 1.
Primer sequences and PCR cycling conditions for amplifying and sequencing full-length pfcrt
| Gene | PCR primer sequence 5′ to 3′ | Cycling conditions | Product size (bp) |
|---|---|---|---|
| pfcrt fragment 1 (exons 1–4) | outer PCR | 94°C3 min, (94°C1 min; 45°C1 min, 65°C1 min) × 30 cycles, 65°C10 min | 873 |
| CRTP1: CCG TTA ATA ATA AAT ACA CGC AG | |||
| NG1_REV: GGA TAC CAT AGC ATT TAA TC | |||
| nested PCR | 94°C3 min, (94°C1 min; 50°C1 min, 65°C1 min) × 30 cycles, 65°C10 min | 827 | |
| CRTP1: CCG TTA ATA ATA AAT ACA CGC AG | |||
| NG4_REV: AGC ATT TAA TCT TAA AAT GTC | |||
| pfcrt fragment 2 (exons 4–13) | outer PCR | 94°C3 min, (94°C1 min; 50°C1 min, 65°C1 min) × 30 cycles, 65°C10 min | 874 |
| NG5_FOR: CTC GGA GCA GTT ATT ATT G | |||
| NG3_Rev: ATT CCT TAT AAA GTG TAA TGC GA | |||
| nested PCR | 94°C3 min, (94°C1 min; 50°C1 min, 65°C1 min) × 30 cycles, 65°C10 min | 827 | |
| NG2_FOR: GCT CTT GTA GAA ATG AAA TTA TC | |||
| NG3_REV: ATT CCT TAT AAA GTG TAA TGC GA | |||
| 5′ and 3′ UTR sequencing primers | NG6_REV: GAA CAT AAT CAT ACA AAT A | ||
| NG7_FOR: CCG TGT AGG AAA TAT TAT CT | |||
| Sequencing primers for confirming intron skipping | P11: TCCTTTTTCCAATTGTTCACTTC | ||
| P13: GACGGAGCATGGGTAAGAAG | |||
| P17: CAAATGGCTTGTTCGTTCATAA |
Results
The full-length cDNA sequence of the pfcrt gene, assembled from two overlapping amplicons together comprising codons 1–424, was successfully obtained from 26 and 35 of the isolates from eastern Sudan and Bagamoyo district, respectively, while codons 1–220 were successfully sequenced in an additional 23 and 13 isolates, respectively.
Expression of an alternatively spliced variant of pfcrt
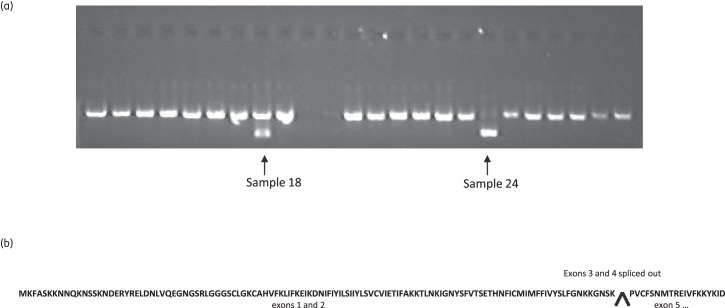
In two isolates from New Halfa, Sudan (samples 18 and 24), the PCR amplifying cDNA from the 5′ segment of the pfcrt mRNA produced two products by gel electrophoresis: the expected 827 bp product and an amplicon of ∼500 bp. In sample 24 this band was more intense than the 827 bp product, while in sample 18 both bands were of moderate intensity (Figure 2a). Direct sequencing of the ∼500 bp PCR product for sample 24 revealed that, for the majority of cDNA molecules amplified, exons 3 and 4 were completely absent, resulting in a truncated cDNA sequence (521 bp; Figure 1b and Figure 2b). This transcript, if translated, encodes a putative 322 amino acid hypothetical protein, which would be 102 amino acids shorter than the canonical PfCRT encoded by the 3D7 clone reference (PF3D7_0709000, www.PlasmoDB.org). It is interesting to note that exons 2 and 5 are both type 0 exons, which means that splicing occurs exactly on codon triplet boundaries (i.e. at the third nucleotide), and so the removal of both exons 3 and 4 does not change the reading phase of the codons. Figure 1(b) illustrates the predicted structure of the full-length spliced cDNA. The sequence of pfcrt mRNA in all remaining patient samples from Gedarif and Fukayosi displayed the expected spliced form. This suggests that the splice variant is not always present.
Figure 2.
(a) Ethidium-bromide-stained agarose gels of RT–PCR products of N-terminus of pfcrt mRNA. Lane 1, Hyper Ladder IV (100 bp); lanes 2–24, clinical samples. Arrows indicate samples 18 and 24 in which two RT–PCR products were observed, the expected 827 bp and the 521 bp truncated product. (b) Predicted amino acid sequence of abnormally spliced pfcrt mRNA from patient 24. The amino acid sequence of exons 1 and 2 is shown. The arrow indicates the splice site at the end of exon 2, followed by the sequence at the beginning of exon 5.
Intron deletion in genomic copies of pfcrt
In order to investigate whether the observed splicing reflects rearrangements at the genomic level, and to address whether pfcrt introns are structured differently in these samples, DNA was extracted from glass fibre membranes by the Chelex method, which does not preserve RNA, from samples 18 and 24, and from three additional randomly selected samples for comparison, 22, 5 and 75 from New Halfa. These were subjected to amplification and sequencing, which was successful only for the 3′ region of the pfcrt gene, coding for the protein carboxy-terminus. Interestingly, amplification produced two PCR products of unexpected sizes: a 600 bp product and an ∼450 bp product in samples 18 and 24, and also in one of the comparators, 22.
For each of these isolates, the two PCR products were excised from agarose gels following electrophoresis, purified and sequenced. Four primers (NG2_FOR, P11, P13 and P17) located in this region were employed for sequencing these products. Direct sequencing of the 600 bp PCR product revealed complete removal of introns, with the analysed partial genomic sequence found to be identical to the predicted mRNA sequence. Figure 3 shows the partial PfCRT amino acid sequence encoded by an intronless gDNA segment amplified from samples 18 and 24. Interpretable sequence data were not obtained from the 450 bp amplicon in any of these isolates. Thus some P. falciparum isolates from New Halfa harbour additional genome copies of pfcrt that lack introns. We were not able to demonstrate that these unusual genomic pfcrt loci were the origin of the unusual transcripts identified in the previous cDNA analysis, and these may be inactive, processed pseudogenes that are not expressed. Alternatively, these may be transcribed by blood stage parasites, giving rise to the unconventionally spliced transcripts observed.
Figure 3.
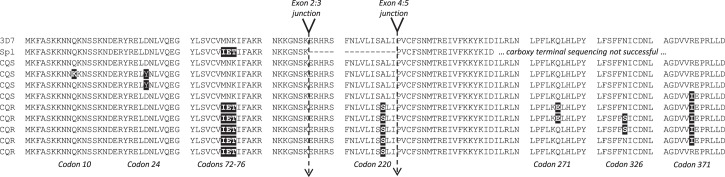
Clustal alignment of predicted amino acid sequences of nine distinct pfcrt haplotypes identified by sequencing cDNA generated directly from peripheral blood parasite mRNA. Contributing data were derived from two New Halfa isolates with identical splice variant transcripts (Sp1), 26 full-length cDNA sequences from New Halfa and Gedaref, Sudan, and 35 full-length cDNA sequences from Fukayosi, Tanzania. Haplotype labels indicate predicted chloroquine resistance status (CQS, susceptible; CQR, resistant). Vertical gaps in the alignment indicate invariant sequence tracts not included. Labels under the alignment refer to variant positions (shaded).
Haplotypes deduced from pfcrt transcript sequences
cDNA sequence data were used to construct extended haplotypes for the pfcrt locus in each P. falciparum isolate that exhibited the wild-type splicing pattern. Two new polymorphisms were observed in a single isolate from Sudan: the non-synonymous nucleotide change C28A generating a Gln to Lys mutation at codon 10 (Q10K), and the nucleotide change G70T resulting in an Asp to Tyr mutation at codon 24 (D24Y). This codon 24 polymorphism was also observed in three isolates from Tanzania. Using the single-letter amino acid nomenclature, the following non-synonymous pfcrt mutations were observed in addition to codons 10 and 24: M74I, N75E, K76T, A220S, Q271E, N326S and R371I. (In the following paragraphs, only these variant positions are considered.)
Among the 23 Sudanese isolates where only the 5′-region of the cDNA was fully sequenced, the wild-type haplotype MNKA (at codons 74–76 and 220) was present in 4 (17.4%), and the remaining 19 isolates carried the mutant IETS haplotype, associated with chloroquine resistance in Africa. Of the 26 isolates for which full-length transcript sequences were obtained, chloroquine-resistant haplotypes dominated, with five distinct full-length haplotypes (Figure 3). A single isolate displayed the reference (wild-type) haplotype MNKAQNR at codons 74–76, 220, 271, 326 and 371. Importantly, two individuals in Sudan harboured parasites with hybrid haplotypes, IETSQNR and IETSQNI, indicative of intragenic recombination within the pfcrt locus in this population.
Among the 13 isolates from Fukayosi, Tanzania, for which only the 5′-region was sequenced, the wild-type MNKA at codons 74–76 and 220 was the predominant allele, being present in 7 isolates (53.8%), with 3 (23.1%) carrying the IETS haplotype. Three patients (23.1%) had a mixture of both the IET and MNK, all of whom carried the mutant 220S. A total of 35 isolates were successfully sequenced for the full-length transcript (Figure 3). Two hybrid haplotypes, IETSQNR and MNKAQNI at codons 74–76, 220, 271, 326 and 371, were detected in a single Tanzanian isolate each. Thus in both Sudan and Tanzania, recombinant pfcrt alleles comprising sequence elements of both chloroquine-resistant and wild-type pfcrt were observed. The prevalence of each haplotype observed in both Sudan and Tanzania is presented in Figure 4.
Figure 4.
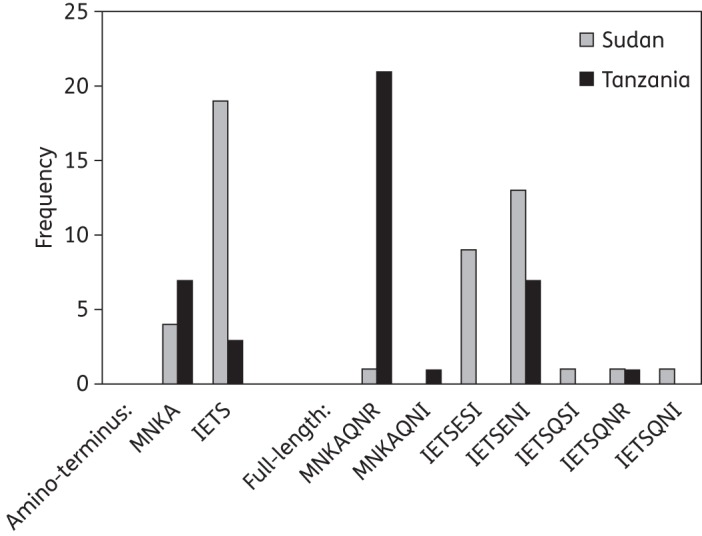
Frequency of pfcrt haplotypes in cDNA from isolates in eastern Sudan (n = 49) and Tanzania (n = 40). Number of observations of each haplotype defined at codons 74–76 and 220 (amino-terminus and full-length data) and 271, 326 and 371 (full-length data only). Data from isolates with multiple genotypes at the pfcrt locus were not included.
Discussion
The CRT membrane transporter is an important modulator of parasite response to antimalarial chemotherapy, and yet few studies have addressed the structure or abundance of pfcrt transcripts in natural P. falciparum infections. Here, we sequenced cDNA amplified from pfcrt mRNA present in P. falciparum-infected peripheral blood samples taken prior to treatment from malaria patients in eastern Sudan and coastal Tanzania. This approach provided evidence that, in vivo, peripheral blood parasites express alternatively spliced pfcrt mRNA, the first such demonstration either in vivo or in vitro, and that naturally occurring parasite genomes can harbour additional pfcrt copies in the form of spliced pseudogenes. Although these extra gene copies may be the source of the unorthodox pfcrt mRNA molecules we detected, additional studies are needed to confirm a direct relationship. Further, we determined extended pfcrt haplotypes, identifying new polymorphisms at codons 10 and 24, and providing the first evidence that chloroquine-resistant and chloroquine-susceptible forms of pfcrt recombine in natural parasite populations.
The unusual variant transcript of pfcrt described here was detected in clinical isolates from only one of two study sites in eastern Sudan, and did not occur among the sequenced Tanzanian isolates. Due to the complete removal of exons 3 and 4 (amino acids 121–222), this transcript encodes a significantly altered putative CRT. This shorter protein will not include the transmembrane domains 3–5, while having a drastically truncated transmembrane domain 6 (Figure 5). Also, this change obliterates three polymorphic amino acid positions (T152A, S163R and the canonical S220A), each probably relevant to CRT function, as they have been associated with different responses of the parasite to several drugs, including chloroquine.10,18 However, we have no evidence to date that such a CRT protein is expressed from the alternatively spliced mRNA, and all isolates expressing this variant transcript also expressed the normally spliced, full-length pfcrt transcript. As exons 2 and 5 are spliced precisely in-frame with codon boundaries (known as type 0 splicing), the downstream sequence frame is maintained, such that the alternative splicing event can generate mRNA molecules that encode in-frame putative polypeptides. This also raised the question as to whether the alternative spliced format of pfcrt and the full-length mRNA were both being expressed from the established pfcrt locus on chromosome 7 (exemplified by PF3D7_0709000).
Figure 5.
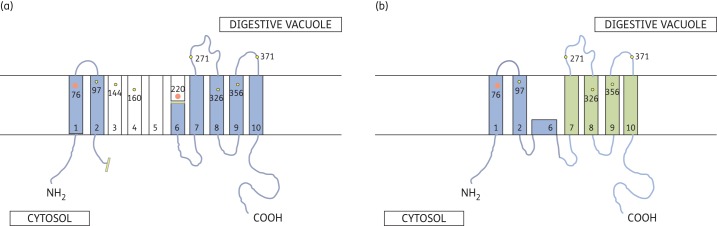
A possible linear arrangement of transmembrane regions in the full-length CRT protein (a) and putative truncated CRT protein encoded by alternative-spliced pfcrt mRNA from Sudanese malaria patients (b). Transmembrane regions are depicted as rectangles; blue denotes presence in putative truncated form of CRT and white denotes presence only in full-length CRT. Freeform blue line denotes extramembrane polypeptide tracts in the cytosol or inside the digestive vacuole lumen. Transverse yellow bars denote site of splicing event and loss of contiguous nucleotide/amino acid sequence. Polymorphic residues are depicted by yellow or orange (codons 76 and 220) dots. Amino terminal polymorphisms at codons 10 and 24 are not shown.
Alternative splicing is an important mechanism by which precursor mRNAs are transcribed into several forms of mature mRNAs,19,20 leading to multiple polypeptides encoded by a single gene, and the generation of plurifunctional variant protein isoforms.21 The first report of a spliced variant mRNA in P. falciparum was the 41-3 gene encoding a variable antigen involved in immune regulation and sought as a vaccine candidate.22 Further examples are the guanylyl cyclase gene involved in the signalling pathway of male gametogenesis,23 the erythrocyte-binding protein MAEBL24 and a CDK-related protein kinase 6.25 Recent transcriptomic studies have identified further variable splicing events for various P. falciparum genes in laboratory-cultured clones such as 3D7 and NF54,26,27 but there have been no reports of alternative splicing in the pfcrt gene in laboratory lines or in vivo. Interestingly, Otto et al.28 described four new splicing events on chromosome 7, but none of these encompassed the pfcrt locus.
In attempting to understand the genomic origin of the variant cDNA sequences we observed, which corresponded to the 5′ half of the normal pfcrt mRNA, the 3′ half of an additional genomic pfcrt locus in which all introns were missing was identified in parasite gDNA amplified from dried filter-paper blood spots. To our knowledge, this is the first description of pseudogenes derived from drug resistance-associated functional genes in Plasmodium. A plausible explanation for our observations, which we were unable to confirm with the material available, is that the intron-less pfcrt locus we have described is a classic pseudogene that has arisen in this parasite population, and is transcribed, as P. falciparum pseudogenes frequently are.28 As such it may have no significant effect on parasite responses to antimalarial drugs, despite being expressed in peripheral blood parasites, although an effect on regulation of transcription from the native pfcrt locus cannot be ruled out. The unusual cDNA we sequenced encompassed the region of pfcrt around the codon 76 polymorphism, suggesting the same may be true for the additional pfcrt pseudogene locus; this is therefore very likely to contribute to genotyping analysis of parasite DNA from peripheral blood. It remains possible that such loci are widespread, and have been part of the ‘signal’ obtained whenever field samples are genotyped at this locus, whether by PCR-RFLP, qPCR or direct sequencing.11,15,29 It is very important that this possible source of confounding of genotyping studies is investigated thoroughly. Interestingly, the pseudogene encodes the mutant 76Thr allele, and so represents an evolutionary event that occurred since chloroquine and related aminoquinolones came into use in the 1940s. It is fascinating to speculate that, as the codon 76 mutation becomes less prevalent due to the withdrawal of chloroquine in Africa, these pseudogenes will remain as genomic ‘fossils’, which may nevertheless provide a source of the mutant sequence that could be reactivated by recombination should chloroquine pressure be resumed in the future. In this context, it should be noted that evidence suggests that considerable variability was present in the pfcrt locus before the era of chloroquine exposure.30
CRT codons 72–76 are currently under opposite selection pressure by artemisinins and partner drugs currently employed in ACT.8,31 This pressure for parasite survival may select specific adaptive changes in gene structure, as well as changes in the regulation and expression of pfcrt and other key loci such as pfmdr1.32–34 This adaptation may include the loss of introns, as reported for fission yeast and mice,35 and the potential gain of new functions. In fact, evidence is gathering that a pseudogene can actually have critical functions, and in some cancers can regulate the ‘parent’ locus.36,37 Finally, our data describe two new polymorphic codons, for amino acids 10 and 24, and provide the first evidence of recombination among chloroquine-resistant and chloroquine-susceptible haplotypes of pfcrt in both Sudan and Tanzania. Such events may indicate that pfcrt genes are still evolving in African parasite populations, generating further heterogeneity in the structure of the CRT protein, and this diversity is likely to be under active selection by ongoing ACT pressure across the continent.
Conclusions
In this study we describe aberrant spliced pfcrt transcripts and confirm the presence of pfcrt pseudogenes in a small number of patient parasite isolates. We also provide evidence from both Sudan and Tanzania that hybrid forms of pfcrt with sequence characteristics of both chloroquine-resistant and wild-type alleles are present in natural populations, probably due to meiotic recombination. A weakness of our study is that the unusual transcripts and alleles of pfcrt were detected in non-replenishable parasite RNA and DNA, and so confirmatory analyses were not possible. Similarly, no phenotypic analysis of any of the parasite isolates was carried out. However, as the variant alleles of pfcrt were identified in more than one individual, and from both mRNA and gDNA sequences, we are confident that these parasites were present in New Halfa. Further studies of pfcrt cDNA sequences from patient peripheral blood are needed to explore this phenomenon further.
Funding
This work was funded by the EU FP7 MALACTRES Consortium, SIDA-SAREC Grant ref. SWE-2009-165 and EU FP7 NanoMal Consortium Grant ref. 304948. N. B. G. was supported by TDR and C. J. S. is supported by Public Health England.
Transparency declarations
None to declare.
References
- 1.White NJ, Nosten F, Looareesuwan S, et al. Averting a malaria disaster. Lancet. 1999;353:1965–7. doi: 10.1016/s0140-6736(98)07367-x. [DOI] [PubMed] [Google Scholar]
- 2.Breman JG. Resistance to artemisinin-based combination therapy. Lancet Infect Dis. 2012;12:820–2. doi: 10.1016/S1473-3099(12)70226-8. [DOI] [PubMed] [Google Scholar]
- 3.Fairhurst RM, Nayyar GM, Breman JG, et al. Artemisinin-resistant malaria: research challenges, opportunities, and public health implications. Am J Trop Med Hyg. 2012;87:231–41. doi: 10.4269/ajtmh.2012.12-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Noedl H, Se Y, Schaecher K, et al. Evidence of artemisinin-resistant malaria in western Cambodia. New Engl J Med. 2008;359:2619–20. doi: 10.1056/NEJMc0805011. [DOI] [PubMed] [Google Scholar]
- 5.Dondorp AM, Nosten F, Yi P, et al. Artemisinin resistance in Plasmodium falciparum malaria. New Engl J Med. 2009;361:455–67. doi: 10.1056/NEJMoa0808859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sisowath C, Stromberg J, Mårtensson A, et al. In vivo selection of Plasmodium falciparum pfmdr1 86N coding alleles by artemether-lumefantrine (Coartem) J Infect Dis. 2005;191:1014–7. doi: 10.1086/427997. [DOI] [PubMed] [Google Scholar]
- 7.Sisowath C, Petersen I, Veiga MI, et al. In vivo selection of Plasmodium falciparum parasites carrying the chloroquine-susceptible pfcrt K76 allele after treatment with artemether-lumefantrine in Africa. J Infect Dis. 2009;199:750–7. doi: 10.1086/596738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Humphreys GA, Merinopoulos I, Ahmed J, et al. Amodiaquine and artemether-lumefantrine select distinct alleles of the Plasmodium falciparum pfmdr1 gene in Tanzanian children treated for uncomplicated malaria. Antimicrob Agents Chemother. 2007;51:991–7. doi: 10.1128/AAC.00875-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gadalla NB, Adam I, Elzaki SE, et al. Increased pfmdr1 copy number and sequence polymorphisms in Plasmodium falciparum isolates from Sudanese malaria patients treated with artemether-lumefantrine. Antimicrob Agents Chemother. 2011;55:5408–11. doi: 10.1128/AAC.05102-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Djimde A, Duombo OK, Cortese JF, et al. A molecular marker for chloroquine-resistant falciparum malaria. New Eng J Med. 2001;344:257–301. doi: 10.1056/NEJM200101253440403. [DOI] [PubMed] [Google Scholar]
- 11.Djimde A, Doumbo OK, Steketee RW, et al. Application of a molecular marker for surveillance of chloroquine-resistant falciparum malaria. Lancet. 2001;358:890–1. doi: 10.1016/S0140-6736(01)06040-8. [DOI] [PubMed] [Google Scholar]
- 12.Sá JM, Twu O, Hayton K, et al. Geographic patterns of Plasmodium falciparum drug resistance distinguished by differential responses to amodiaquine and chloroquine. Proc Natl Acad Sci USA. 2009;106:18883–9. doi: 10.1073/pnas.0911317106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beshir K, Sutherland CJ, Merinopoulos I, et al. Amodiaquine resistance in Plasmodium falciparum malaria is associated with the pfcrt 72–76 SVMNT allele in Afghanistan. Antimicrob Agents Chemother. 2010;54:3714–6. doi: 10.1128/AAC.00358-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Waller KL, Muhle RA, Ursos LM, et al. Chloroquine resistance modulated in vitro by expression levels of the Plasmodium falciparum chloroquine resistance transporter. J Biol Chem. 2003;278:33593–601. doi: 10.1074/jbc.M302215200. [DOI] [PubMed] [Google Scholar]
- 15.Chen N, Kyle DE, Pasay C, et al. pfcrt allelic types with two novel amino acid mutations in chloroquine-resistant Plasmodium falciparum isolates from the Philippines. Antimicrob Agents Chemother. 2003;47:3500–5. doi: 10.1128/AAC.47.11.3500-3505.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hietala SF, Mårtensson A, Ngasala B, et al. Population pharmacokinetics and pharmacodynamics of artemether and lumefantrine during combination treatment in children with uncomplicated falciparum malaria in Tanzania. Antimicrob Agents Chemother. 2010;54:4780–8. doi: 10.1128/AAC.00252-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carlsson AM, Ngasala BE, Dahlstrom S, et al. Plasmodium falciparum population dynamics during the early phase of anti-malarial drug treatment in Tanzanian children with acute uncomplicated malaria. Malar J. 2011;10:380. doi: 10.1186/1475-2875-10-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson DJ, Fidock DA, Mungthin M, et al. Evidence for a central role for PfCRT in conferring Plasmodium falciparum resistance to diverse antimalarial agents. Mol Cell. 2004;15:867–77. doi: 10.1016/j.molcel.2004.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lopez AJ. Alternative splicing of pre-mRNA: developmental consequences and mechanisms of regulation. Annu Rev Genet. 1998;32:279–305. doi: 10.1146/annurev.genet.32.1.279. [DOI] [PubMed] [Google Scholar]
- 20.Smith CW, Valcarcel J. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem Sci. 2000;25:381–8. doi: 10.1016/s0968-0004(00)01604-2. [DOI] [PubMed] [Google Scholar]
- 21.Gamazon ER, Stranger BE. Genomics of alternative splicing: evolution, development and pathophysiology. Hum Genet. 2014;133:679–87. doi: 10.1007/s00439-013-1411-3. [DOI] [PubMed] [Google Scholar]
- 22.Knapp B, Nau U, Hundt E, et al. Demonstration of alternative splicing of a pre-mRNA expressed in the blood stage form of Plasmodium falciparum. J Biol Chem. 1991;266:7148–54. [PubMed] [Google Scholar]
- 23.Carucci DJ, Witney AA, Muhia DK, et al. Guanylyl cyclase activity associated with putative bifunctional integral membrane proteins in Plasmodium falciparum. J Biol Chem. 2000;275:22147–56. doi: 10.1074/jbc.M001021200. [DOI] [PubMed] [Google Scholar]
- 24.Singh N, Preiser P, Rénia L, et al. Conservation and developmental control of alternative splicing in maebl among malaria parasites. J Mol Biol. 2004;343:589–99. doi: 10.1016/j.jmb.2004.08.047. [DOI] [PubMed] [Google Scholar]
- 25.Bracchi-Ricard V, Barik S, Delvecchio C, et al. PfPK6, a novel cyclin-dependent kinase/mitogen-activated protein kinase-related protein kinase from Plasmodium falciparum. Biochem J. 2000;347:255–63. [PMC free article] [PubMed] [Google Scholar]
- 26.Iriko H, Jin L, Kaneko O, et al. A small-scale systematic analysis of alternative splicing in Plasmodium falciparum. Parasitol Int. 2009;58:196–9. doi: 10.1016/j.parint.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 27.van Dooren GG, Su V, D'Ombrain MC, et al. Processing of an apicoplast leader sequence in Plasmodium falciparum and the identification of a putative leader cleavage enzyme. J Biol Chem. 2002;277:23612–9. doi: 10.1074/jbc.M201748200. [DOI] [PubMed] [Google Scholar]
- 28.Otto TD, Wilinski D, Assefa S, et al. New insights into the blood-stage transcriptome of Plasmodium falciparum using RNA-Seq. Mol Microbiol. 2010;76:12–24. doi: 10.1111/j.1365-2958.2009.07026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gadalla NB, Elzaki SE, Mukhtar E, et al. Dynamics of pfcrt alleles CVMNK and CVIET in chloroquine-treated Sudanese patients infected with Plasmodium falciparum. Malar J. 2010;9:74. doi: 10.1186/1475-2875-9-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chan CW, Spathis R, Reiff DM, et al. Diversity of Plasmodium falciparum chloroquine resistance transporter (pfcrt) exon 2 haplotypes in the Pacific from 1959 to 1979. PLoS One. 2012;7:e30213. doi: 10.1371/journal.pone.0030213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malmberg M, Ngasala B, Ferreira PE, et al. Temporal trends of molecular markers associated with artemether-lumefantrine tolerance/resistance in Bagamoyo district, Tanzania. Malar J. 2013;12:103. doi: 10.1186/1475-2875-12-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Veiga MI, Ferreira PE, Jörnhagen L, et al. Novel polymorphisms in Plasmodium falciparum ABC transporter genes are associated with major ACT antimalarial drug resistance. PLoS One. 2011;6:e20212. doi: 10.1371/journal.pone.0020212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Price RN, Uhlemann AC, Brockman A, et al. Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet. 2004;364:438–47. doi: 10.1016/S0140-6736(04)16767-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sidhu AB, Uhlemann AC, Valderramos SG, et al. Decreasing pfmdr1 copy number in Plasmodium falciparum malaria heightens susceptibility to mefloquine, lumefantrine, halofantrine, quinine, and artemisinin. J Infect Dis. 2006;194:528–35. doi: 10.1086/507115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jeffares DC, Penkett CJ, Bahler J. Rapidly regulated genes are intron poor. Trends Genet. 2008;24:375–8. doi: 10.1016/j.tig.2008.05.006. [DOI] [PubMed] [Google Scholar]
- 36.Li W, Yang W, Wang XJ. Pseudogenes: pseudo or real functional elements? J Genet Genomics. 2013;40:171–7. doi: 10.1016/j.jgg.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 37.Wang L, Guo ZY, Zhang R, et al. Pseudogene OCT4-pg4 functions as a natural micro RNA sponge to regulate OCT4 expression by competing for miR-145 in hepatocellular carcinoma. Carcinogenesis. 2013;34:1773–81. doi: 10.1093/carcin/bgt139. [DOI] [PubMed] [Google Scholar]