Figure 4.
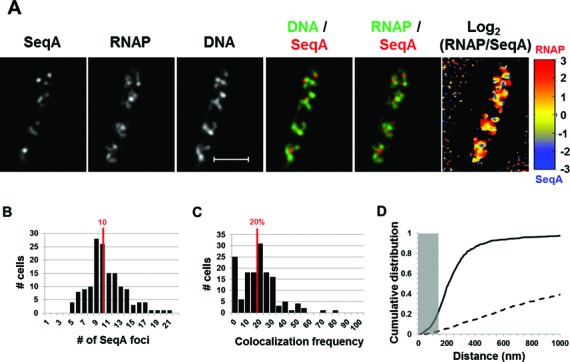
Spatial segregation of transcription foci and replication forks tracked by SeqA in fast-growing cells. (A) Images of SeqA, RNAP, DNA (nucleoid), overlays of SeqA (red) and DNA (green), RNAP (green) and SeqA (red) and log2 (RNAP/SeqA) (heat map) from a representative fast-growing E. coli cell as described in the legend to Figure 1. SeqA foci are largely located at different positions from transcription foci (red and green colors on the overlay of SeqA and RNAP and red and blue color on the heat map). Note also that SeqA foci appear mainly to be separated from high intensities of DNA signals in the nucleoids (red and green colors on the overlay of SeqA and DNA). (B) The distribution of apparent SeqA-mCherry foci in a population of fast-growing cells. The red line in the histogram indicates the median number of SeqA foci in those cells. (C) Co-localization frequency between the SeqA foci and transcription foci in fast-growing cells. The red line indicates the median co-localization frequency of SeqA foci with transcription foci in the population of cells as described in the Materials and Methods section. Majority of SeqA foci are separated from transcription foci in the fast-growing cells. The average co-localization frequency of SeqA foci and RNAP foci from a random data set is 0%. (D) Cumulative distribution of the distances between SeqA foci and their closest RNAP foci in the population of cells as described in the Materials and Methods section. (──) SeqA-mCherry RNAP-Venus and (- - -) SeqA-mCherry RNAP-Venus random. The gray rectangle represents the co-localization area (≤140 nm). Only 21.5% of the SeqA foci are within 140 nm of the closest transcription foci.
