Figure 1.
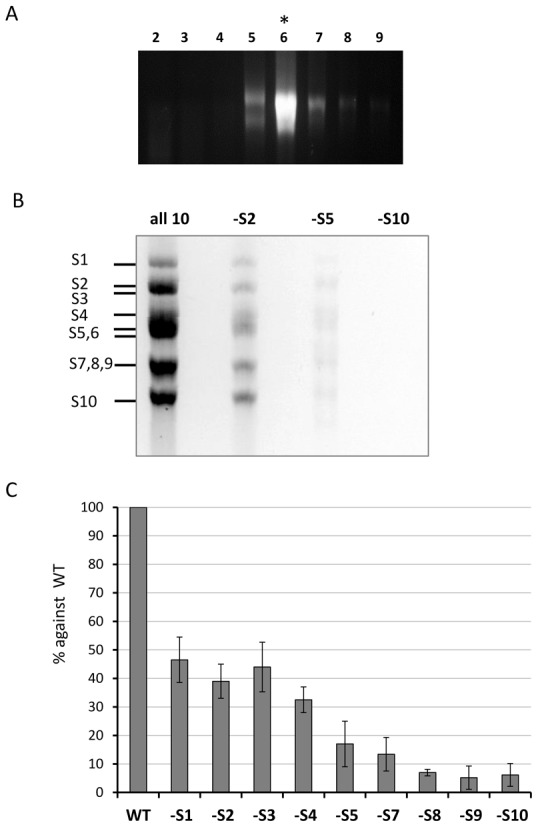
Exclusion of specific BTV RNA segment influences genome packaging. An incomplete set of 32P-labelled BTV ssRNAs that excludes S2 or S5 or S10 (indicated as -S2, -S5, -S10) but includes all the 9 respective segments were used in the in vitro CFA assay; the reaction mixture was purified on a sucrose gradient. A complete set of 10 ssRNAs was also included (all 10) as a control. (A) RNA distribution within the sucrose gradient fractions 2–9 was analysed on an agarose gel. The fraction containing assembled cores (fraction 6) is indicated with an asterix (*). (B) Packaged RNA profile after RNAse digestion and purification of fraction 6 analysed by 1% denaturing agarose gel. Segments S1–S10 are indicated on the left. (C) Quantification of the effect of segment exclusion. Ten BTV ssRNAs (WT) or ssRNAs excluding one ssRNA at a time (-S1, -S2, etc.) were used in the CFA assay. Packaged ssRNA in relevant fraction containing cores was purified on a sucrose gradient. BTV segment S6 was quantified by qRT-PCR to represent the packaging efficiency. Quantities of S6 in samples of -S1, -S2, etc. were compared with WT control in the same experiment and packaging efficiency was calculated. Standard deviations from three independent experiments are indicated (error bars).
