Figure 1.
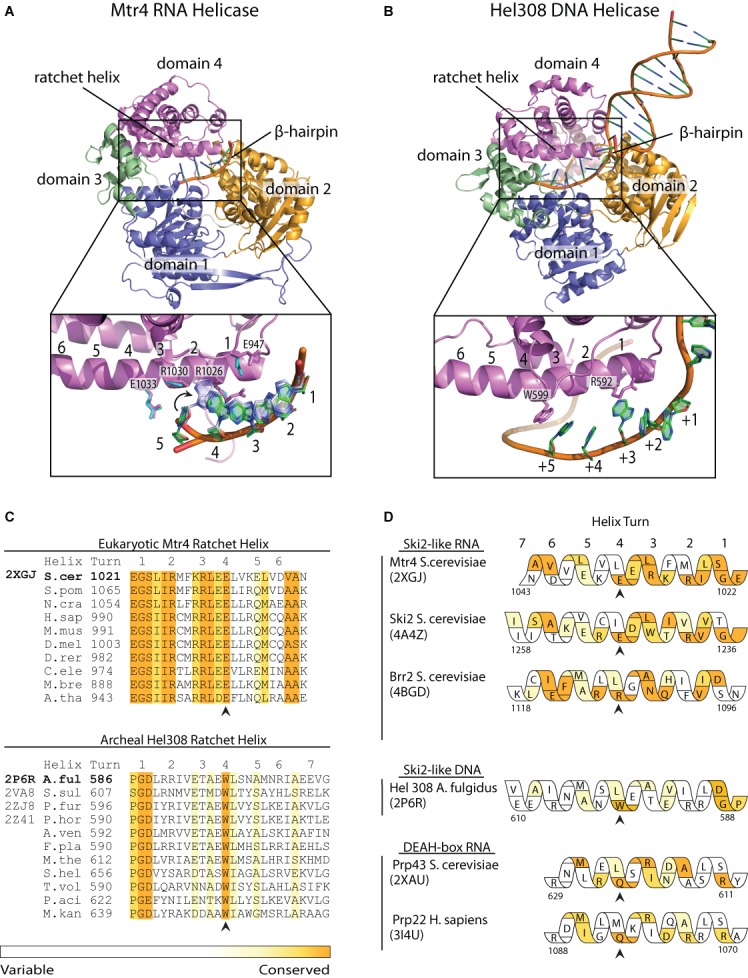
Conserved ratchet helix residues interact with nucleic acid. (A) The RNA-bound Mtr4 structure (PDB 2XGJ) is colored by domain. (Inset) Helix bundle domain (domain 4 or ratchet domain) residues that interact with poly(A) RNA are shown as sticks. For reference, the ratchet helix is numbered by helical turn in an N- to C-terminal direction. Aligned residues (cyan) and RNA bases (light blue) from a second molecule in the asymmetric unit are also indicated. Nucleotides are labeled 1 through 5 in a 5′-3′ direction. (B) The DNA-bound Hel308 structure (PDB 2P6R) is colored as in (A). The inset image highlights the pi stacking interactions of ratchet helix residues W599 and R592 shown as sticks with bases of ssDNA. Nucleotides downstream of the strand separation β-hairpin are labeled +1 through +5. (C) Alignment and conservation scores (calculated using Consurf (50)) of eukaryotic Mtr4 and archaeal Hel308 ratchet helix sequences. Conservation is colored strictly conserved as orange, to variable as white. Extensive conservation at helical turn 4 is highlighted with an arrow. Alignment of Mtr4 includes 10 model eukaryotic species (S.cer, Saccharomyces cerevisiae; S.pom, Schizosaccharomyces pombe; N.cra, Neurospora crassa; H.sap, Homo sapiens; M.mus, Mus musculus; D.rer, Danio rerio; D.mel, Drosophila melanogaster; C.ele, Caenorhabditis elegans; M.bre, Monosiga brevicollis; A.tha, Arabidopsis thaliana). Alignment of Hel308 includes the sequences of crystal structure homologs and seven other archeal sequences (A.ful, Archaeoglobus fulgidus; S.sul, Sulfolobus solfataricus; P.fur, Pyrococcus furiosus; P.hor, Pyrococcushorikoshi; A.ven, Archaeoglobus fulgidus; F. pla, Ferroglobus placidus; M.the, Methanosaeta thermophila S.hel, Staphylothermus hellenicus; T.vol, Thermoplasma volcanium P.aci, Candidatus Parvarchaeum acidophilus; M.kan, Methanopyrus kandleri). (D) Conservation of each residue along the ratchet helix of Ski2-like and DEAH/RHA-box helicases is depicted with Consurf scores colored as in (C). Complete sequence alignments are shown in Supplementary Figure S2. Conservation at helical turn 4 is highlighted with an arrow. Structures in (A) and (B) were rendered using PyMOL (62).
