Figure 3.
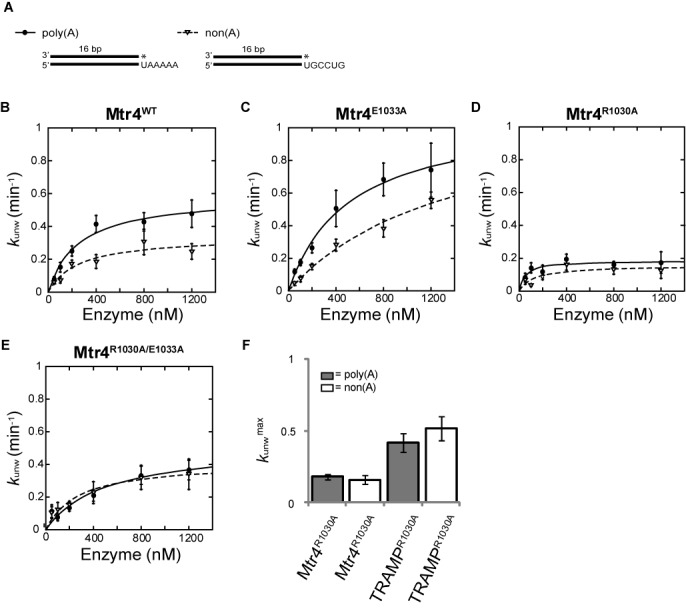
Role for R1030 in 3′ poly(A) sequence recognition. (A) Comparison of the poly(A) and non(A) RNA substrates used in this study. Each substrate is composed of identical 16 bp duplex regions and variable 3′ end overhangs (see ‘Materials and Methods’ for full sequence details). (B) The Mtr4R1030A mutant loses the ability to discriminate between a poly(A) and non(A) RNA substrate. Unwinding rate (kunw) constants determined for Mtr4WT (A) and Mtr4 mutants (B, C, D, E), plotted as a function of Mtr4 concentration using poly(A) (solid line) and non(A) (dashed line) RNA substrates. Although Mtr4R1030A/E1033A restores unwinding activity to wild-type levels, no difference is observed between poly(A) and non(A) substrates (E). Best fit curves to the data were calculated as described in ‘Materials and Methods’. Data presented here represent averages from three independent experiments; error bars represent SD. (F) Comparison of kunwmax values for Mtr4 and TRAMP complexes containing the R1030A mutation (Mtr4R1030A and TRAMPR1030A) shows that TRAMPR1030A also loses the ability to discriminate between poly(A) and non(A) RNA substrates. Data presented here represent averages from four independent experiments; error bars represent SD.
