Table 1. Kinetic parameters (kunwmax and K1/2) of Mtr4 mutants for unwinding poly(A) and non(A) substratesa.
| poly(A) | non(A) | poly(A) | non(A) | |
|---|---|---|---|---|
| Enzyme | kunwmax (min−1) | kunwmax (min−1) |
K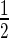 (nM) (nM) |
K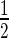 (nM) (nM) |
| Mtr4WT | 0.59 ± 0.05 | 0.34 ± 0.05 | 252 ± 60 | 255 ± 116 |
| Mtr4archless | 0.49 ± 0.07 | 0.34 ± 0.08 | 221 ± 93 | 260 ± 164 |
| Mtr4R1030A | 0.18 ± 0.02 | 0.16 ± 0.03 | 51 ± 26 | 129 ± 92 |
| Mtr4E1033A | 1.08 ± 0.09b | 1.17 ± 0.24b | 504 ± 93 | 1415 ± 468 |
| Mtr4E1033W | 0.22 ± 0.07 | undetermined | 484 ± 360 | undetermined |
| Mtr4R1030A/E1033A | 0.52 ± 0.1 | 0.41 ± 0.04 | 498 ± 216 | 269 ± 81 |
| Mtr4R1030A-archless | n.d. | n.d. | n.d. | n.d. |
| Mtr4E1033A-archless | n.d. | n.d. | n.d. | n.d. |
Data presented here represent averages from three independent experiments; error bars represent SD. n.d., no unwinding activity detected.
aMethodologies and equations used to derive kinetic constants are found in the ‘Materials and Methods’ section.
bSince the curve for the Mtr4E1033A catalyzed unwinding never reached saturation, calculated kunwmax rates between poly(A) and non(A) appear within error, although a clear distinction between RNA substrates is observed at every tested concentration (see Figure 3).
