Figure 4.
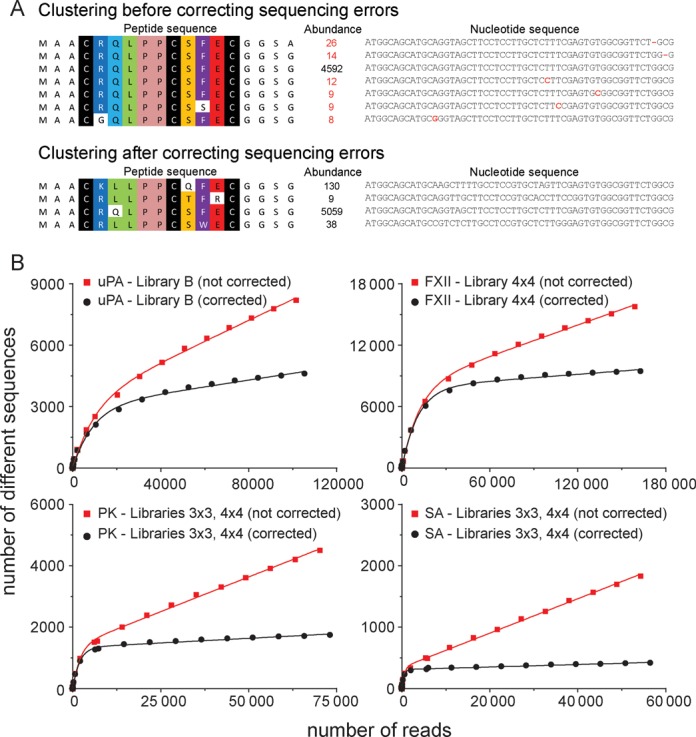
(A) Example for the identification of false consensus sequences due to sequencing errors. In a selection of Library A against SrtA, the most abundant sequence (present 4592 times) was clustered with sequences differing in only 1 nucleotide and being present at much lower frequency. These sequences likely resulted from sequencing errors. A MatLab script (fixingerrors.m script) was developed to eliminate these erroneous sequences. For the high-abundance sequence shown in the figure, 467 erroneous sequences were found (9%). For other high-abundance sequences, wrong sequences ranged between 0 and 48%. (B) Examples of saturation plots for different datasets before and after correcting sequencing errors (all datasets were obtained after one round of phage selection).
