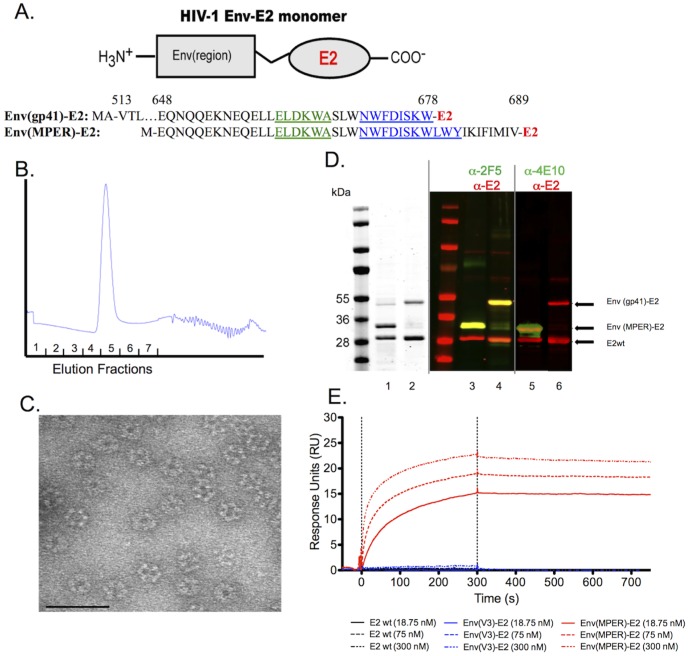
Figure 1. Construction and antigenic characterization of the HIV-1 Env(MPER)-E2 and Env(gp41)-E2 particles.
(A) Schematic illustration of an E2 monomer with the HIV-1 Env region fused to the N-terminus. Amino acid sequences of the Env(MPER)-E2 and Env(gp41)-E2 constructs are shown below and numerated in relationship to the reference strain HXB2. Within the amino acid sequences, the 2F5 mAb epitope is underlined and highlighted in green and the 4E10 mAb epitope is underlined and highlighted in blue. (B) Representative SuperdexX200 gel filtration chromatograph of purified Env(MPER)-E2 particles. (C) Representative transmission electron micrograph of HIV-1 Env(gp41)-E2 particles. Env(MPER)-E2 and E2wt particles were of similar size and shape. Scale bar is 75 nm. (D) SimplyBlue stain of Env(MPER)-E2 (lane 1) and Env(gp41)-E2 (lane 2) revealing the purity of these particles at 93% and 90%, respectively. Western blot analysis of Env(MPER)-E2 (lanes 3 and 5) and Env(gp41)-E2 (lanes 4 and 6) using the Li-COR Odyssey dual-detection system to demonstrate the identity of these particles. Red bands indicate proteins detected with anti-E2 Abs, and green bands indicate proteins detected by either 2F5 or 4E10 mAbs. Yellow bands indicate proteins that are detected by both anti-E2 and either 2F5 or 4E10 mAbs. (E) Surface Plasmon Resonance Spectroscopy of Env(MPER)-E2 particles binding to the 2F5 NmAb to demonstrate accessibility of the 2F5 epitope on the surface of the scaffold. Binding was detected by flowing 2F5 mAb concentrations of 18.75 nM, 75 nM and 300 nM over E2 particles immobilized on a CM3 chip. No binding of the E2wt scaffold (lacking HIV fusion proteins) or Env(V3)-E2 particles to the 2F5 mAb were detected.