Figures 1 and 3 are incorrect. Please view the corrected Figures 1 and 3, as well as the corrected corresponding figure legends, here.
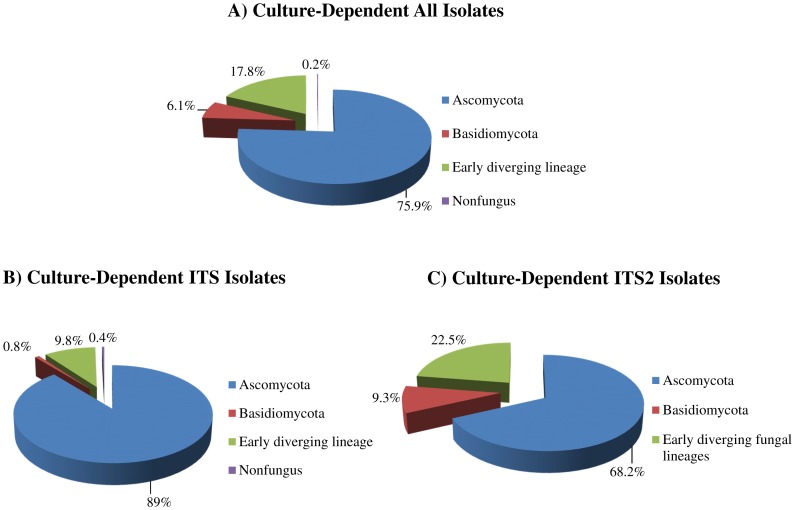
Figure 1. Fungal isolates recovered by culture-dependent methods.
(A) Relative distribution of all cultures according to different fungal phyla; (B) Relative distribution of isolates identified by homologies to ITS sequences; (C) Relative distribution of isolates identified by homologies to ITS2 sequences.
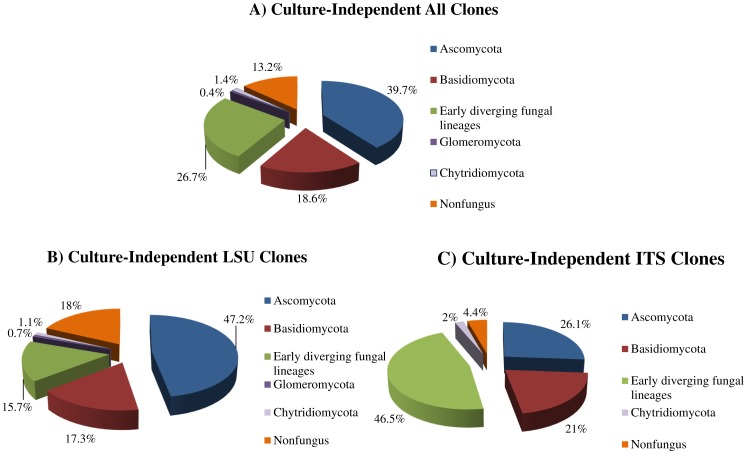
Figure 3. Phyla distribution in culture-independent (CI) clones:
A) Relative proportions of different phyla of all clones; B) Relative proportions of different phyla of LSU clones; C) Relative proportions of different phyla of ITS clones.
Reference
- 1. Zhang T, Victor TR, Rajkumar SS, Li X, Okoniewski JC, et al. (2014) Mycobiome of the Bat White Nose Syndrome Affected Caves and Mines Reveals Diversity of Fungi and Local Adaptation by the Fungal Pathogen Pseudogymnoascus(Geomyces) destructans . PLoS ONE 9(9): e108714 doi:10.1371/journal.pone.0108714 [DOI] [PMC free article] [PubMed] [Google Scholar]