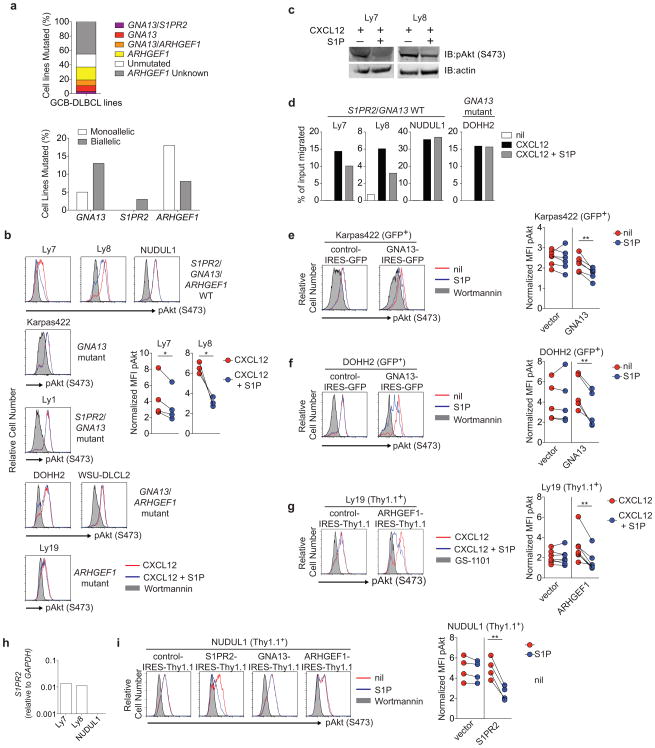
Extended Data Figure 5. Defective regulation of pAkt and cell migration in human GCB DLBCL cell lines harboring mutations in the S1PR2 signaling pathway.
(a) Frequency of non-synonymous coding mutations in S1PR2, GNA13 and ARHGEF1 in GCB-DLBCL lines, and fraction that were mono- or bi-allelic, summarized from Supplementary Table 3. Unmutated indicates no coding region mutations in the genes shown. (b, c) Intracellular FACS (b) or Western blot (c) for pAkt in human GCB DLBCL cell lines that are WT or mutant for S1PR2, GNA13 or ARHGEF1 as indicated and that were stimulated with CXCL12 (100 ng/ml) in the presence or absence of S1P (10 nM) for 5 minutes. pAkt staining of cells treated with wortmannin (200 nM) for 5 minutes are shown in gray as a staining control for each cell line. (d) Transwell migration of GNA13 WT (Ly7, Ly8, NUDUL1) or mutant (DOHH2) cell lines to CXCL12 (100 ng/ml) in the presence or absence of S1P (10 nM). (e, f) Intracellular FACS for pAkt of the GNA13 mutant cell lines Karpas422 (d) or DOHH (e) transduced with retrovirus expressing the reporter alone (vector) or GNA13 in the presence or absence of S1P (10 nM) or wortmannin (200 nM; staining control). (g) Intracellular FACS for pAkt in the ARHGEF1 mutant cell line Ly19 transduced with retrovirus expressing reporter alone (vector) or ARHGEF1 that were treated as in b or with the PI3K inhibitor GS-1101 (2 uM; staining control). (h) Quantitative PCR analysis of S1PR2 transcript abundance in human GCB-DLBCL cell lines relative to GAPDH. (i) Intracellular FACS for pAkt in NUDUL1 cells transduced with retrovirus expressing reporter alone (vector), S1PR2, GNA13 or ARHGEF1, treated as in d. Data in b and d are representative of at least 3 independent experiments. Pooled data from at least 3 independent experiments is shown in b, e, f, g and i. Data in b is one experiment representative of 2. **P<0.01, paired two-tailed Student's t test.