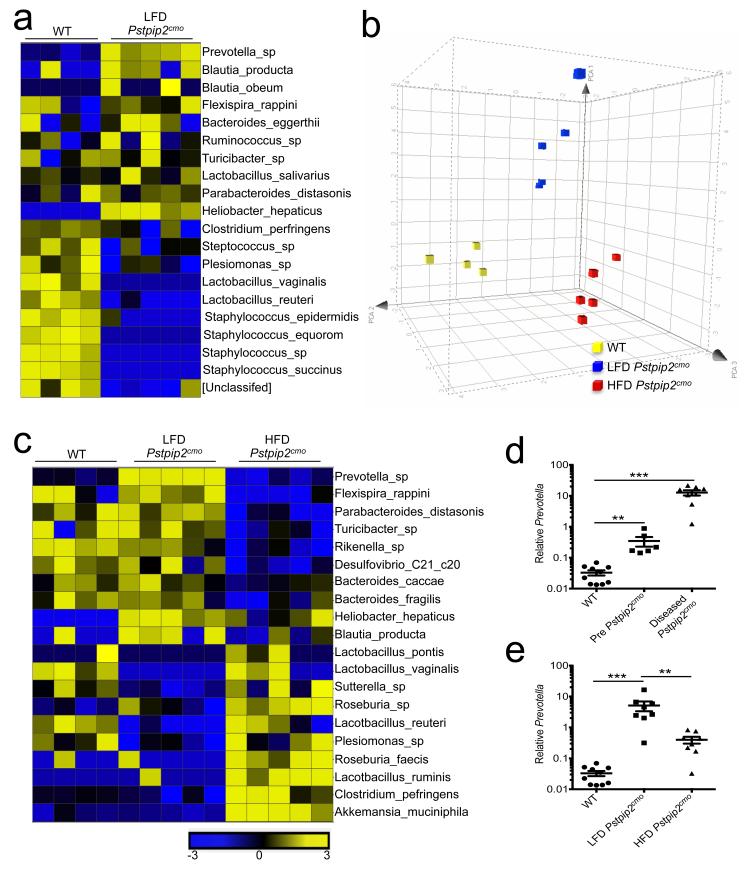
Figure 2. Alterations in commensal microbiota landscape that are associated with Pstpip2cmo-mediated osteomyelitic disease can be modified by changes in diet.
a-c, Fecal samples were collected from WT, LFD Pstpip2cmo and HFD Pstpip2cmo mice at 10-12 weeks of age and 16S rRNA metagenomic sequencing was conducted. a, Heat map of fold differences in relative abundance of commensal bacteria. b, Principal coordinated analysis (PCoA) plot of fecal microbiota. c, Heat map of the top 20 commensal genera and species that differ between LFD Pstpip2cmo and HFD Pstpip2cmo mice are presented. d, Prevotella 16S rDNA copy numbers in WT and Pstpip2cmo mice before (pre-disease: 3-6 weeks of age) and after (diseased: 10-16 weeks of age) the development of osteomyelitis. Each point represents an individual mouse, and the line represents the mean ± s.e.m. Data are representative of four independent experiments. e, 16S rDNA analysis of Prevotella abundance. Data are representative of four independent experiments. (**P < 0.01, ***P < 0.001; Student’s t-test)