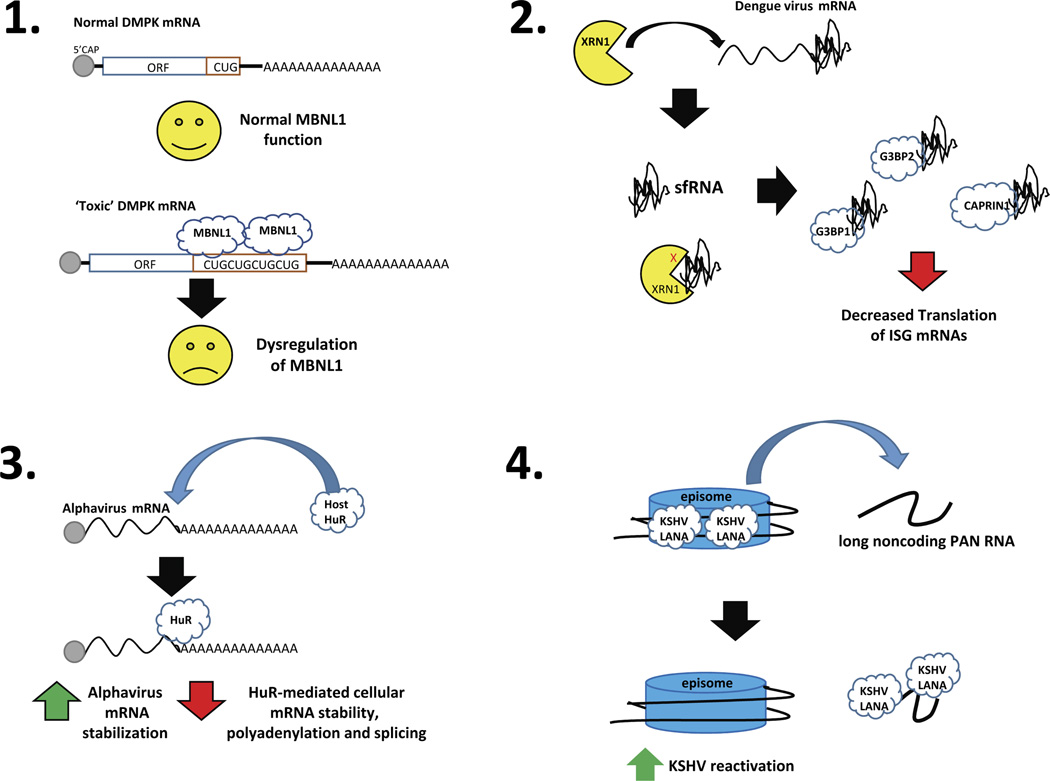
Figure 1.
The functional consequences of four examples of RNA-mediated protein sponging. Panel 1: Expanded CUG repeat sequences in the 3’ UTR of the DMPK mRNA serve as a sponge for the muscleblind (MBNL1) RNA binding protein, resulting in dysregulated RNA splicing that contributes to the disease myotonic dystrophy. Panel 2: Dengue virus mRNA contains a structure in its 3’ UTR that stalls and represses the cellular XRN1 exoribonuclease. The large amount of the stable decay intermediates – called sfRNA – is then able to sponge the indicated cellular RNA binding proteins and repress translation of mRNAs involved in innate immunity. Panel 3: Alphavirus mRNAs contain a high affinity binding site in their 3’ UTRs that binds the cellular HuR protein, resulting in increased viral RNA stability as well as sponging of the cellular factor that dysregulates its normal function. Panel 4: The KSHV PAN RNA serves as a sponge for the viral LANA protein, removing it from viral episomes and facilitating reactivation of latent virus.