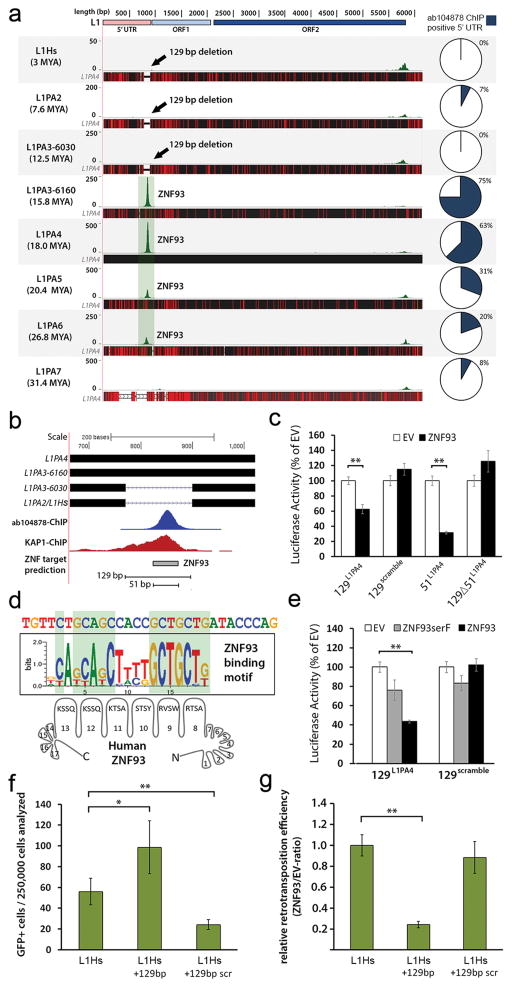
Figure 3. L1PA elements are repressed by primate-specific ZNF93.
a, Green peaks represent genome-wide ab104878-ChIP-seq peak-summits mapped to L1PA consensus sequences. Black horizontal bars: alignment to L1PA4, red lines: divergent positions. b, The 129 bp deletion and predicted 51 bp ZNF93 binding motif (grey bar) relative to L1PA4. c, Relative activity of OCT4-enhancer-luciferase reporters after co-transfection of EV or ZNF93. d, Consensus central sequence of ab104878-ChIP-seq summits for L1PA4, aligned with the predicted recognition motif of ZNF93 zinc fingers 8-13. e, Relative activity for OCT4-enhancer-luciferase reporters after co-transfection of EV, ZNF93SerF or ZNF93. f, Number of GFP-positive cells derived from retrotransposition events of L1Hs, L1Hs+129 and L1Hs+129-scrambled constructs in HEK cells (n=7). g, Same as (f) but showing the ratio of retrotransposition events after co-transfection with ZNF93 compared to EV. Error bars: SEM. *= p<0.05; **= p<0.01