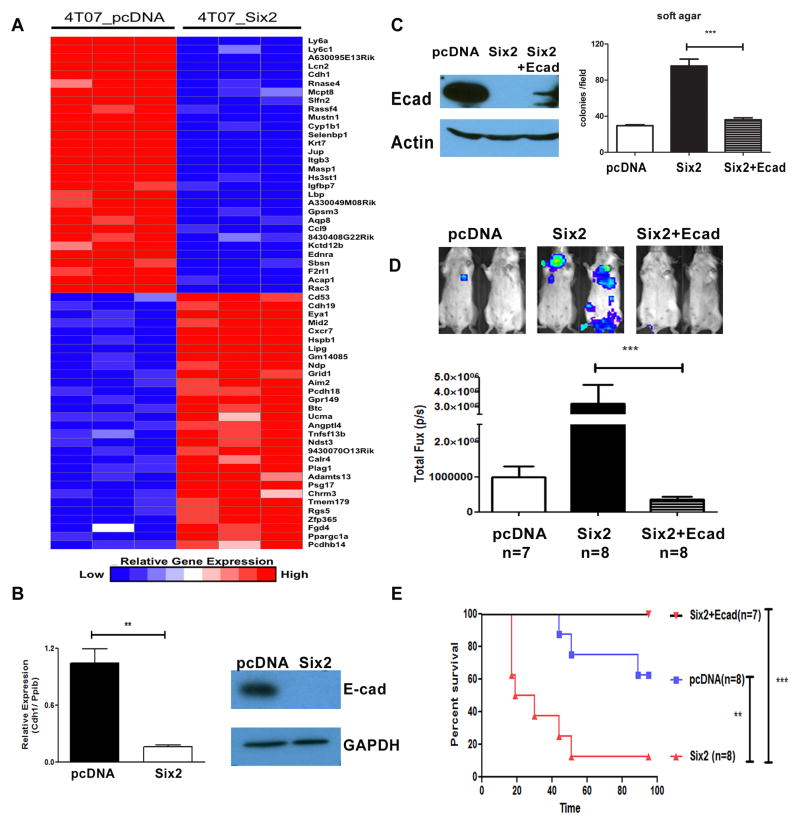
Fig 5. Restoration of E-Cadherin in 4TO7-Six2 cells suppresses Six2-mediated metastasis and increases survival in animals.
(A) Gene expression using microarray analysis from 4TO7-pcDNA and 4TO7-Six2 was examined (each cell line microarray was performed in triplicate). Expression data is shown as top-regulated genes in 4TO7-pcDNA and 4TO7-Six2 cells. The color scale represents the expression level of a gene above (red) and below (blue) the mean expression level of that gene across all samples. (B) E-cadherin mRNA expression was determined by real-time PCR, normalized by Cyclophilin in 4TO7-pcDNA and 4TO7-Six2 cells (left). E-cadherin protein expression was measured by Western blotting. GAPDH was used as loading control (right). (C) E-cadherin expression in 4TO7-pcDNA, Six2, and Six2+Ecad was measured by Western blotting.β-actin was used as loading control (Left). Restoration of E-cadherin in 4TO7-Six2 expressing cells decreases anchorage independent cell growth (Right). Quantification of colony numbers formed by 4TO7-pcDNA, 4TO7-Six2, or 4TO7-Six2+Ecad cells. ***, P<0.001. (D) Luciferase labeled 4TO7-pcDNA, 4TO7-Six2, or 4TO7-Six2+Ecad cells were injected into Balb/c mice through the tail vein and in vivo metastasis was measured using IVIS imaging. Representative pictures from each group are shown (top) and quantification of luciferase signal in each animal injected with 4TO7-pcDNA (n=8), 4TO7-Six2 (n=8), or 4TO7-Six2+Ecad (n=7) is shown (bottom). (E) Kaplan-Meier plot shows overall survival of the injected mice. Statistical analysis was performed using the log-rank test.