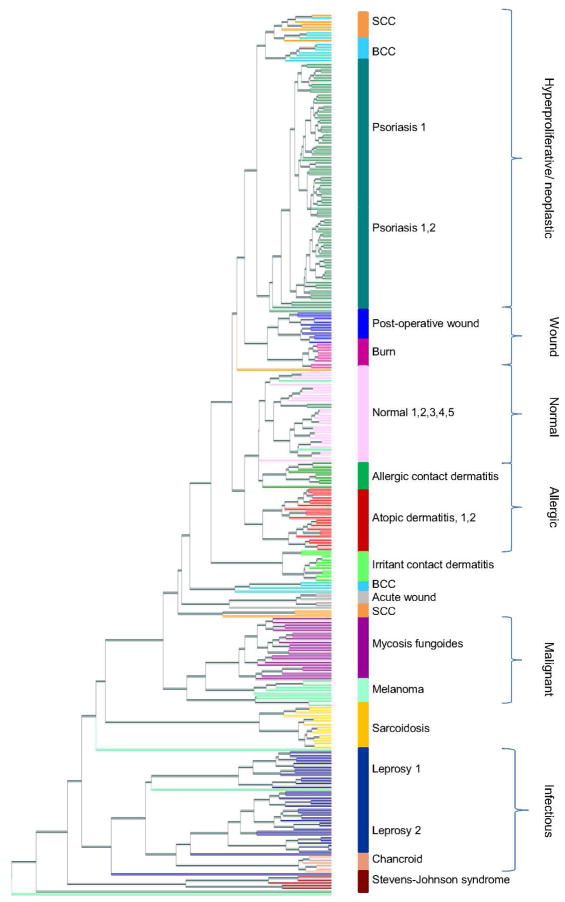
Figure 1. Unsupervised hierarchical clustering tree of 311 skin samples.
Normalized, filtered gene expression profiles were clustered using gene expression distance (Pearson correlation) and displayed in a tree. Each terminal leaf in the tree represents a biopsy sample and is colored according to disease, with colored bars to the right representing the majority disease diagnosis. Samples that clustered apart from other samples of the same diagnosis can be seen a leaf that differs in color from its neighbors. Numbers following disease name labels denote batches of the same disease, and lists of numbers following a disease name denote multiple batches clustering to the same tree branch with little or no differentiation by batch. Brackets to the far right delineate biological groups of neighboring diseases.