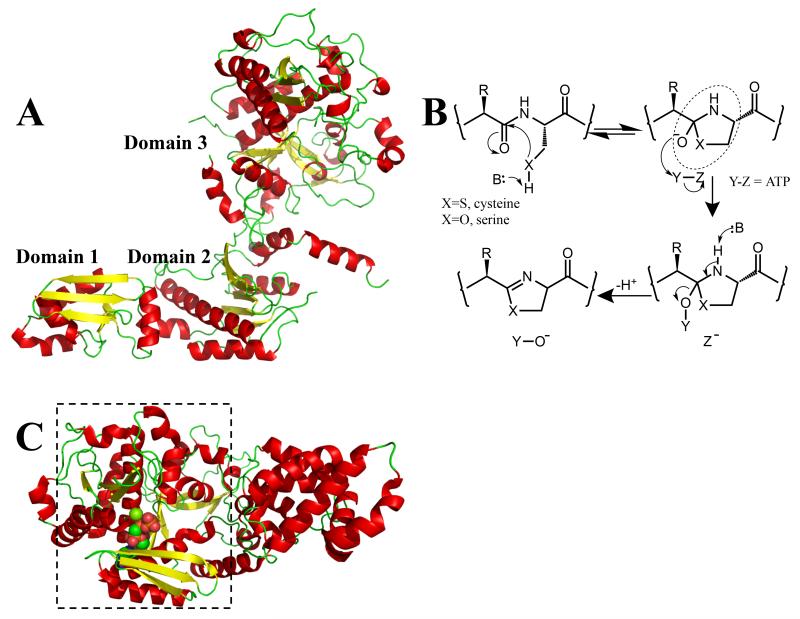
Figure 2.
(a) The structure of TruD (cartoon with α-helices in red; β-sheets in yellow; β-turns in green) shows it has three domains [23••]. Domains 1 and 2 are very similar to a known adenylase MccB [15] and to BalhC from the TOMM pathway. Domain 3 has a novel structure and is related in sequence to BalhD in the TOMM pathway [17••]
(b) heterocyclization of amino acids (cysteine X = S, selenocysteine X = Se, serine/threonine X = O) to yield azoline rings. TOMMS were first predicted to go through a hemiorthamide intermediate (circled). It is likely that in the patellamide biosynthetic pathway, heterocyclization goes through the same central intermediate. In the TOMM system [17••] ATP is used to phosphorylate (Y = ADP, Z = PO43−) the intermediate, whereas in the patellamide pathway the intermediate was proposed to be adenylated (Y = AMP, Z = P2O74−) [23••]. Either mechanism would be irreversible and would promote elimination yielding the azoline. There is a third possibility, pyrophosphorylation (Y = P2O74−, Z = AMP), which although rare, has precedent and is consistent with much of the published data;
(c) a cartoon representation of the structure of the YcaO domain [18••] from E. coli has located the nucleotide binding site. This domain is boxed and is structurally similar to domain 3 of TruD [23••], shown in Figure 2a. The binding of ATP to domain 3 disfavors the adenylase mechanism previously proposed [23••]. The protein is colored as Figure 2a, the nucleotide analog AMP-PNP is shown in space fill with carbon atoms in green, nitrogen atoms in blue and oxygen atoms in red.