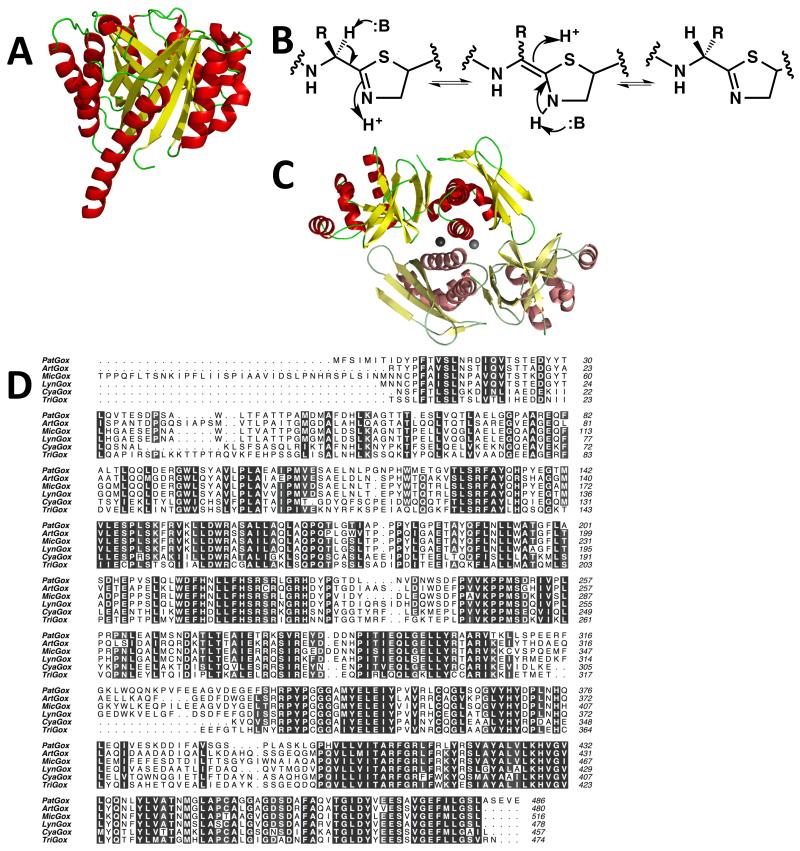
Figure 4.
(a) The structure of PatF [37] represented as Figure 2a. Although the structure is that of a prenyl transferase, the protein is inactive. In other biosynthetic pathways, homologs of PatF have been shown to be active [35 and 36]
(b) a mechanism for the epimerization at the carbon adjacent to a thiazoline. The, study of a chemically related compound has led to the suggestion that in the patellamide pathway this is a spontaneous reaction [40]. However, an enzyme catalyzed processes has not been formally ruled out nor has epimerization adjacent to a thiazole been excluded
(c) cartoon representation of the structure of the PatG C-terminal DUF [39] reveals it to be a dimer, linked by zinc atoms. One monomer is colored as Figure 2a, the second monomer has duller colors
(d) the oxidase domain of PatG is highly conserved in other patellamide-like biosynthetic pathways with PatG homologs. Clustal Omega sequence alignment of the N-terminal oxidase domains of cyanobactin proteins PatG (Prochloron didemini), ArtG (Arthrospira platensis), MicG (Microcystis aeruginosa NIES-298), LynG (Lyngbya sp. PCC 8106), CyaG (Cyanothece sp. PCC7822) and TriG (Trichodesmium erythraeum IMS101). Figure created using ALINE.