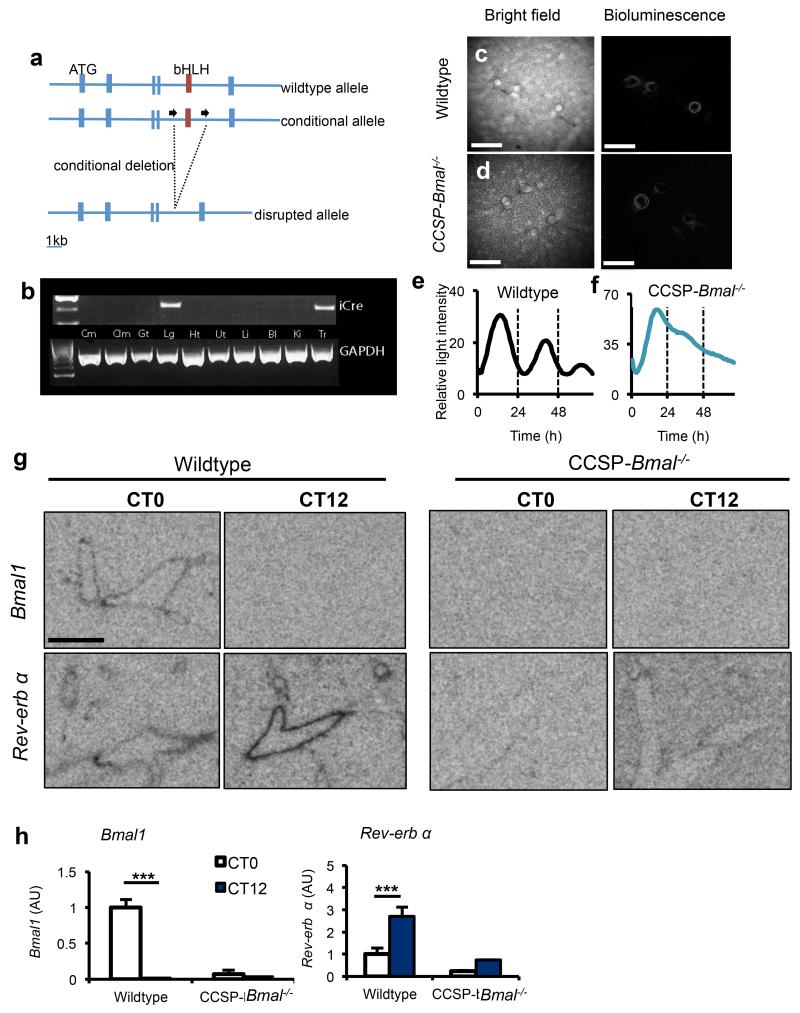
Figure 2.
Targeted ablation of Bmal1 in CCSP expressing cells disrupts circadian rhythmicity in whole lung. (a) Schematic illustrating the region surrounding the basic helix-loop-helix (bHLH) domain of the mouse Bmal1 locus, the conditional floxed allele and the disrupted allele. (b) Expression of iCre (and the housekeeping gene Gapdh) in tissues harvested from CCSP-Bmal−/− (Bmal1fl/fl; CCSP-icre+/−) mice (Cm, cerebrum; Clm, cerebellum; Gt, gut; Lg, lung; Ht, heart; Ut, uterus; Li, liver; Bl, bladder; Ki, kidney; Tr, trachea). (c,d) Ectopic lung slices from Bmal1fl/fl; CCSP-icre+/− (CCSP-Bmal−/−) or CCSP-icre negative littermate controls (wildtype) on a Per2-luc background were placed under a bioluminescence camera. Scale bars, 500 μm. (e,f) Bioluminescence intensity from bronchioles quantified and plotted as a function of time. Data are representative of 3 independent trials. (g, h) Quantification of clock genes Bmal1 and Nr1d1 (rev-erb α) expression in the bronchioles of lungs at CT0 (n=4/genotype) and CT12 (n=3/genotype) (scale bar, 5 mm, two-way ANOVA and post hoc Bonferroni. Data are expressed as mean ± s.e.m and *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.005.