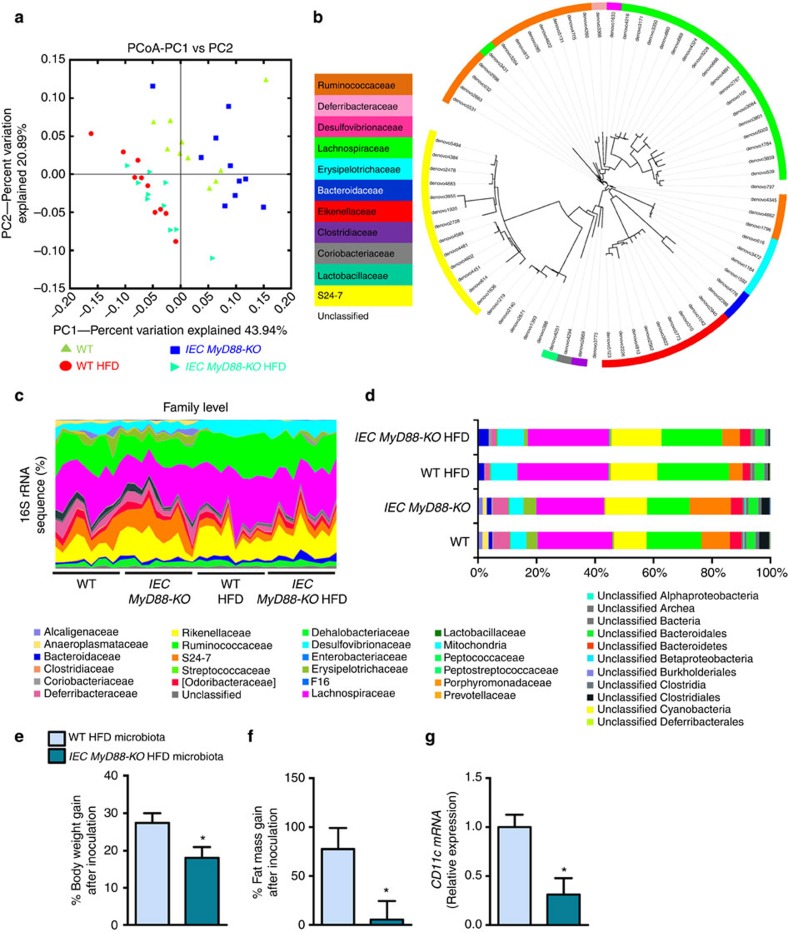
Figure 8. Intestinal MyD88 deletion affects gut bacterial community.
Gut bacterial community is analysed by 16S rRNA gene high-throughput sequencing. IEC MyD88-KO HFD gut microbiota transfers the partial protection against the obesity phenotype to germ-free mice. (a) Principal coordinate analysis based on the weighted UniFrac analysis on operational taxonomic units (OTUs; n=10). Each symbol representing a single sample is coloured according to the group. (b) OTUs significantly affected by intestinal epithelial MyD88 deletion under HFD. A representative 16S rRNA gene from each of the 72 differentially expressed OTUs in WT HFD versus IEC MyD88-KO HFD mice was aligned and used to infer the phylogenetic tree shown in this figure (n=10). The colour in front of the OTU indicates the family of the OTU. (c) Relative abundances (percentage of 16S rRNA gene sequences) of the different bacterial families in each sample among the WT, IEC MyD88-KO, WT HFD and IEC MyD88-KO HFD mice (n=10). (d) Percentage of each indicated family (n=10). The different families are represented by different colour codes. e–g are the results of gut microbiota transfer from WT HFD or IEC MyD88-KO HFD mice to WT germ-free mice fed a HFD. (e) Percentage of body weight gain after gut microbiota transfer to germ-free mice (%; n=5 CT and 4 HFD). (f) Percentage of fat mass gain after gut microbiota transfer to germ-free mice (%; n=5 CT and 4 HFD). (g) CD11c mRNA in the adipose tissue (n=5 CT and 4 HFD). These data correspond to the results of one experiment. All the replicates represent biological replicates. Data are shown as the means±s.e.m. Data with * are significantly different (P<0.05) according to the unpaired two-tailed Student’s t-test.