Figure 2.
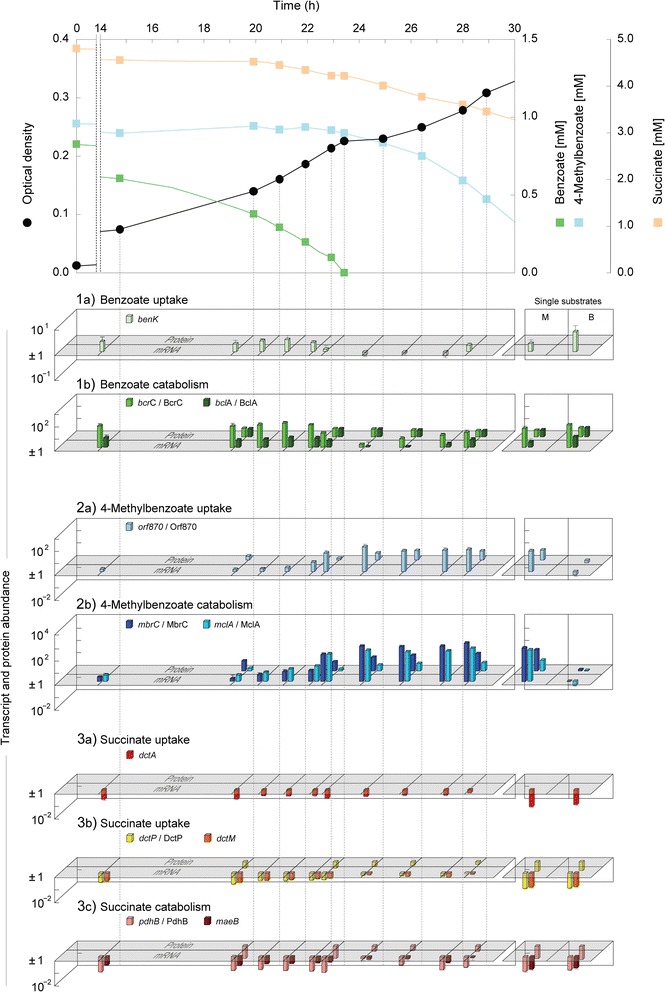
Time-resolved profiles of selected transcripts and corresponding proteins (if identified by 2D DIGE) related to uptake and catabolism of benzoate (1ab), 4-methylbenzoate (2ab) and succinate (3abc). 4-Methylbenzoate-adapted cells of Magnetospirillum sp. strain pMbN1 were shifted to a ternary substrate mixture (see Figure 1d) of 4-methylbenzoate (adaptation substrate), benzoate (co-substrate) and succinate (co-substrate). Abbreviations for experiments with single substrates: M, 4-methylbenzoate; B, benzoate. The fold change in protein and ratio of transcript abundance were determined by comparison to succinate-adapted cells as reference. See legend to Figure 3 for name and predicted functions of selected transcripts/proteins. For detailed information see Additional file 1: Tables S1 and S2.
